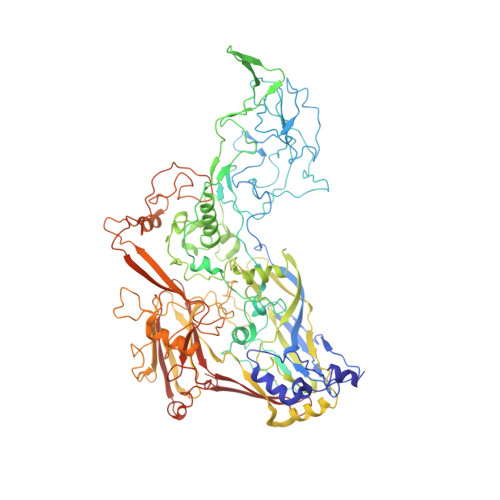
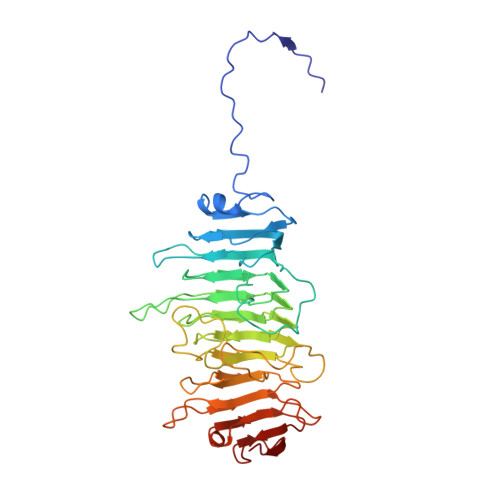
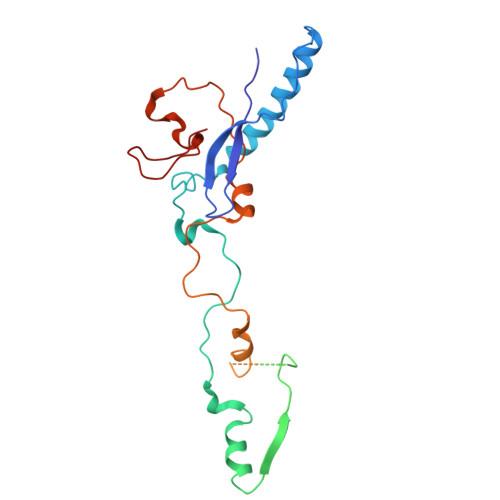
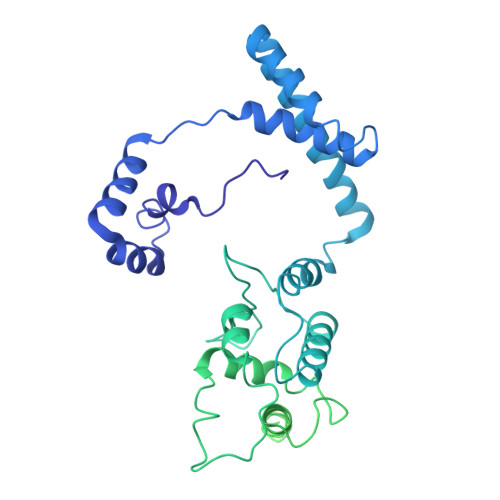
Near-atomic structure of an atadenovirus reveals a conserved capsid-binding motif and intergenera variations in cementing proteins.
Marabini, R., Condezo, G.N., Krupovic, M., Menendez-Conejero, R., Gomez-Blanco, J., San Martin, C.(2021) Sci Adv 7
- PubMed: 33789897
- DOI: https://doi.org/10.1126/sciadv.abe6008
- Primary Citation of Related Structures:
6QI5 - PubMed Abstract:
Of five known adenovirus genera, high-resolution structures are available only for mammalian-infecting mastadenoviruses. We present the first high-resolution structure of an adenovirus with nonmammalian host: lizard atadenovirus LAdV-2. We find a large conformational difference in the internal vertex protein IIIa between mast- and atadenoviruses, induced by the presence of an extended polypeptide. This polypeptide, and α-helical clusters beneath the facet, likely correspond to genus-specific proteins LH2 and p32k. Another genus-specific protein, LH3, with a fold typical of bacteriophage tailspikes, contacts the capsid surface via a triskelion structure identical to that used by mastadenovirus protein IX, revealing a conserved capsid-binding motif and an ancient gene duplication event. Our data also suggest that mastadenovirus E1B-55 K was exapted from the atadenovirus-like LH3 protein. This work provides new information on the evolution of adenoviruses, emphasizing the importance of minor coat proteins for determining specific physicochemical properties of virions and most likely their tropism.
Organizational Affiliation:
Escuela Politécnica Superior, Universidad Autónoma de Madrid, Francisco Tomás y Valiente 11, 28049 Madrid, Spain.