A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Sernee, M.F., Ralton, J.E., Nero, T.L., Sobala, L.F., Kloehn, J., Vieira-Lara, M.A., Cobbold, S.A., Stanton, L., Pires, D.E.V., Hanssen, E., Males, A., Ward, T., Bastidas, L.M., van der Peet, P.L., Parker, M.W., Ascher, D.B., Williams, S.J., Davies, G.J., McConville, M.J.(2019) Cell Host Microbe 26: 385-399.e9
- PubMed: 31513773
- DOI: https://doi.org/10.1016/j.chom.2019.08.009
- Primary Citation of Related Structures:
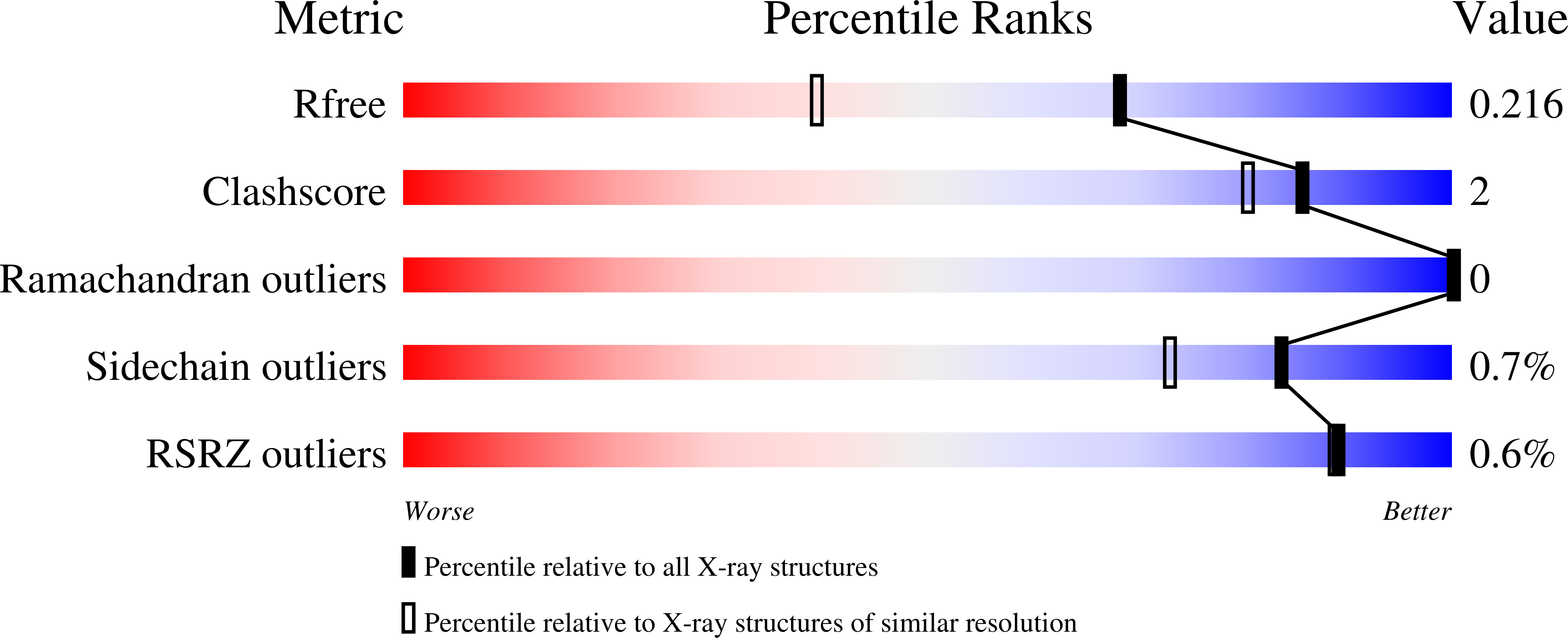
6Q4W, 6Q4X, 6Q4Y, 6Q4Z, 6Q50 - PubMed Abstract:
Parasitic protists belonging to the genus Leishmania synthesize the non-canonical carbohydrate reserve, mannogen, which is composed of β-1,2-mannan oligosaccharides. Here, we identify a class of dual-activity mannosyltransferase/phosphorylases (MTPs) that catalyze both the sugar nucleotide-dependent biosynthesis and phosphorolytic turnover of mannogen. Structural and phylogenic analysis shows that while the MTPs are structurally related to bacterial mannan phosphorylases, they constitute a distinct family of glycosyltransferases (GT108) that have likely been acquired by horizontal gene transfer from gram-positive bacteria. The seven MTPs catalyze the constitutive synthesis and turnover of mannogen. This metabolic rheostat protects obligate intracellular parasite stages from nutrient excess, and is essential for thermotolerance and parasite infectivity in the mammalian host. Our results suggest that the acquisition and expansion of the MTP family in Leishmania increased the metabolic flexibility of these protists and contributed to their capacity to colonize new host niches.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Bio21 Molecular Science and Biotechnology Institute, University of Melbourne, Parkville, VIC 3010, Australia.