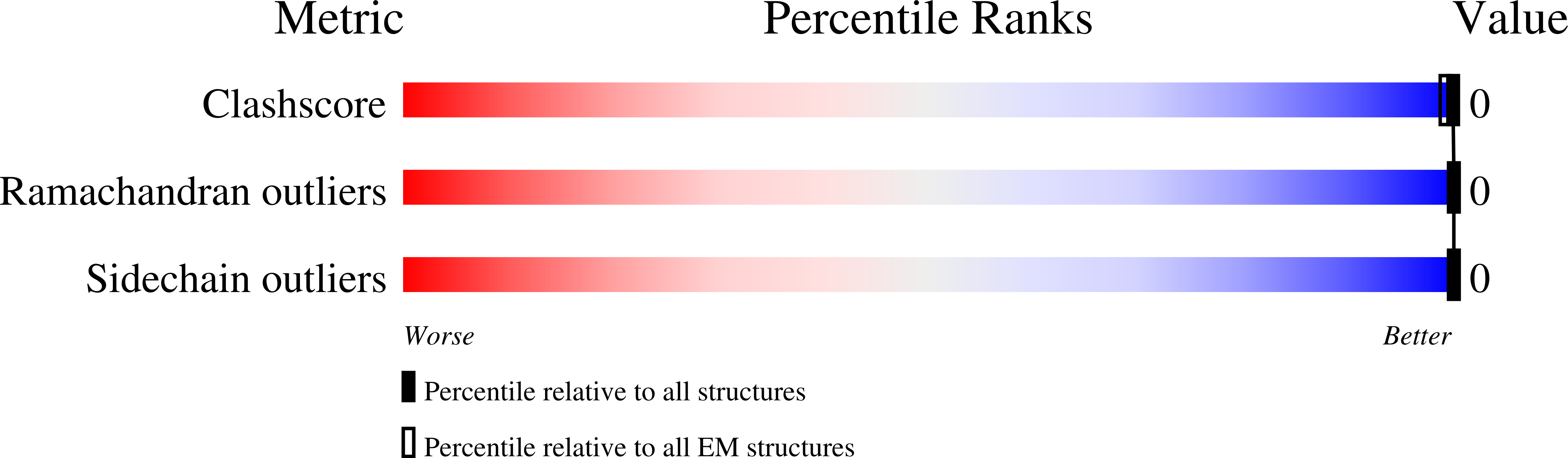Enhancing and shaping the immunogenicity of native-like HIV-1 envelope trimers with a two-component protein nanoparticle.
Brouwer, P.J.M., Antanasijevic, A., Berndsen, Z., Yasmeen, A., Fiala, B., Bijl, T.P.L., Bontjer, I., Bale, J.B., Sheffler, W., Allen, J.D., Schorcht, A., Burger, J.A., Camacho, M., Ellis, D., Cottrell, C.A., Behrens, A.J., Catalano, M., Del Moral-Sanchez, I., Ketas, T.J., LaBranche, C., van Gils, M.J., Sliepen, K., Stewart, L.J., Crispin, M., Montefiori, D.C., Baker, D., Moore, J.P., Klasse, P.J., Ward, A.B., King, N.P., Sanders, R.W.(2019) Nat Commun 10: 4272-4272
- PubMed: 31537780
- DOI: https://doi.org/10.1038/s41467-019-12080-1
- Primary Citation of Related Structures:
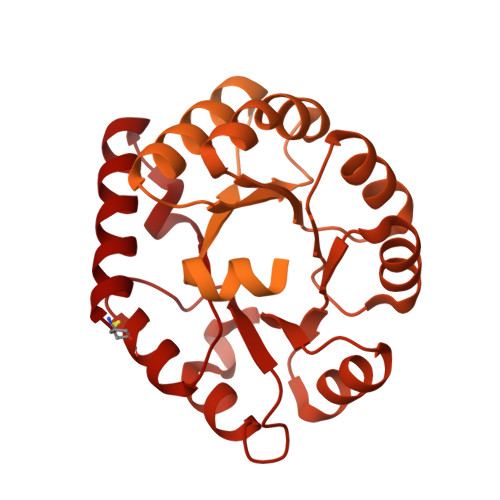
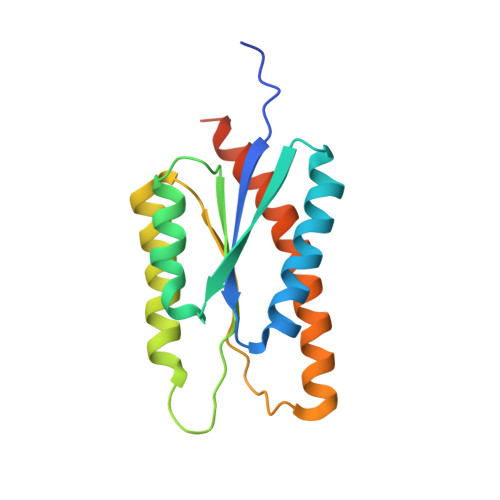
6P6F - PubMed Abstract:
The development of native-like HIV-1 envelope (Env) trimer antigens has enabled the induction of neutralizing antibody (NAb) responses against neutralization-resistant HIV-1 strains in animal models. However, NAb responses are relatively weak and narrow in specificity. Displaying antigens in a multivalent fashion on nanoparticles (NPs) is an established strategy to increase their immunogenicity. Here we present the design and characterization of two-component protein NPs displaying 20 stabilized SOSIP trimers from various HIV-1 strains. The two-component nature permits the incorporation of exclusively well-folded, native-like Env trimers into NPs that self-assemble in vitro with high efficiency. Immunization studies show that the NPs are particularly efficacious as priming immunogens, improve the quality of the Ab response over a conventional one-component nanoparticle system, and are most effective when SOSIP trimers with an apex-proximate neutralizing epitope are displayed. Their ability to enhance and shape the immunogenicity of SOSIP trimers make these NPs a promising immunogen platform.
Organizational Affiliation:
Amsterdam UMC, Department of Medical Microbiology, Amsterdam Infection & Immunity Institute, University of Amsterdam, Amsterdam, 1105AZ, The Netherlands.