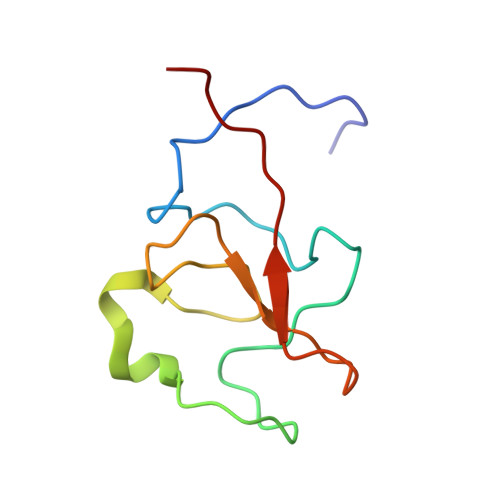
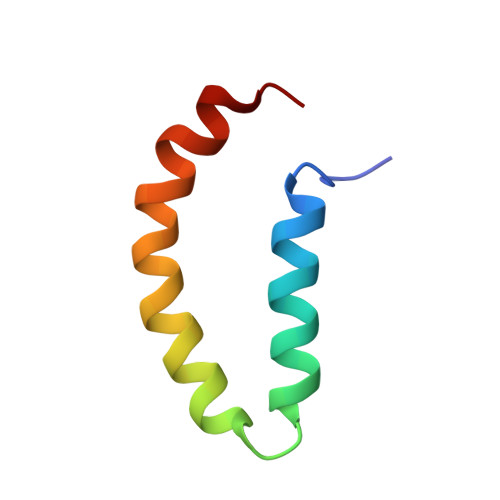
Solution structural model of the complex of the binding regions of human plasminogen with its M-protein receptor from Streptococcus pyogenes.
Yuan, Y., Ayinuola, Y.A., Singh, D., Ayinuola, O., Mayfield, J.A., Quek, A., Whisstock, J.C., Law, R.H.P., Lee, S.W., Ploplis, V.A., Castellino, F.J.(2019) J Struct Biol 208: 18-29
- PubMed: 31301349
- DOI: https://doi.org/10.1016/j.jsb.2019.07.005
- Primary Citation of Related Structures:
6OKW, 6OKX, 6OKY, 6OQ9, 6OQJ, 6OQK - PubMed Abstract:
VEK50 is a truncated peptide from a Streptococcal pyogenes surface human plasminogen (hPg) binding M-protein (PAM). VEK50 contains the full A-domain of PAM, which is responsible for its low nanomolar binding to hPg. The interaction of VEK50 with kringle 2, the PAM-binding domain in hPg (K2 hPg ), has been studied by high-resolution NMR spectroscopy. The data show that each VEK50 monomer in solution contains two tight binding sites for K2 hPg , one each in the a1- (RH1; R 17 H 18 ) and a2- (RH2; R 30 H 31 ) repeats within the A-domain of VEK50. Two mutant forms of VEK50, viz., VEK50[RH1/AA] (VEK50 ΔRH1 ) and VEK50[RH2/AA] (VEK50 ΔRH2 ), were designed by replacing each RH with AA, thus eliminating one of the K2 hPg binding sites within VEK50, and allowing separate study of each binding site. Using 13 C- and 15 N-labeled peptides, NMR-derived solution structures of VEK50 in its complex with K2 hPg were solved. We conclude that the A-domain of PAM can accommodate two molecules of K2 hPg docked within a short distance of each other, and the strength of the binding is slightly different for each site. The solution structure of the VEK50/K2 hPg , complex, which is a reductionist model of the PAM/hPg complex, provides insights for the binding mechanism of PAM to a host protein, a process that is critical to S. pyogenes virulence.
Organizational Affiliation:
W.M. Keck Center for Transgene Research, University of Notre Dame, Notre Dame, IN 46556, USA.