MORC3 Is a Target of the Influenza A Viral Protein NS1.
Zhang, Y., Ahn, J., Green, K.J., Vann, K.R., Black, J., Brooke, C.B., Kutateladze, T.G.(2019) Structure 27: 1029-1033.e3
- PubMed: 31006586
- DOI: https://doi.org/10.1016/j.str.2019.03.015
- Primary Citation of Related Structures:
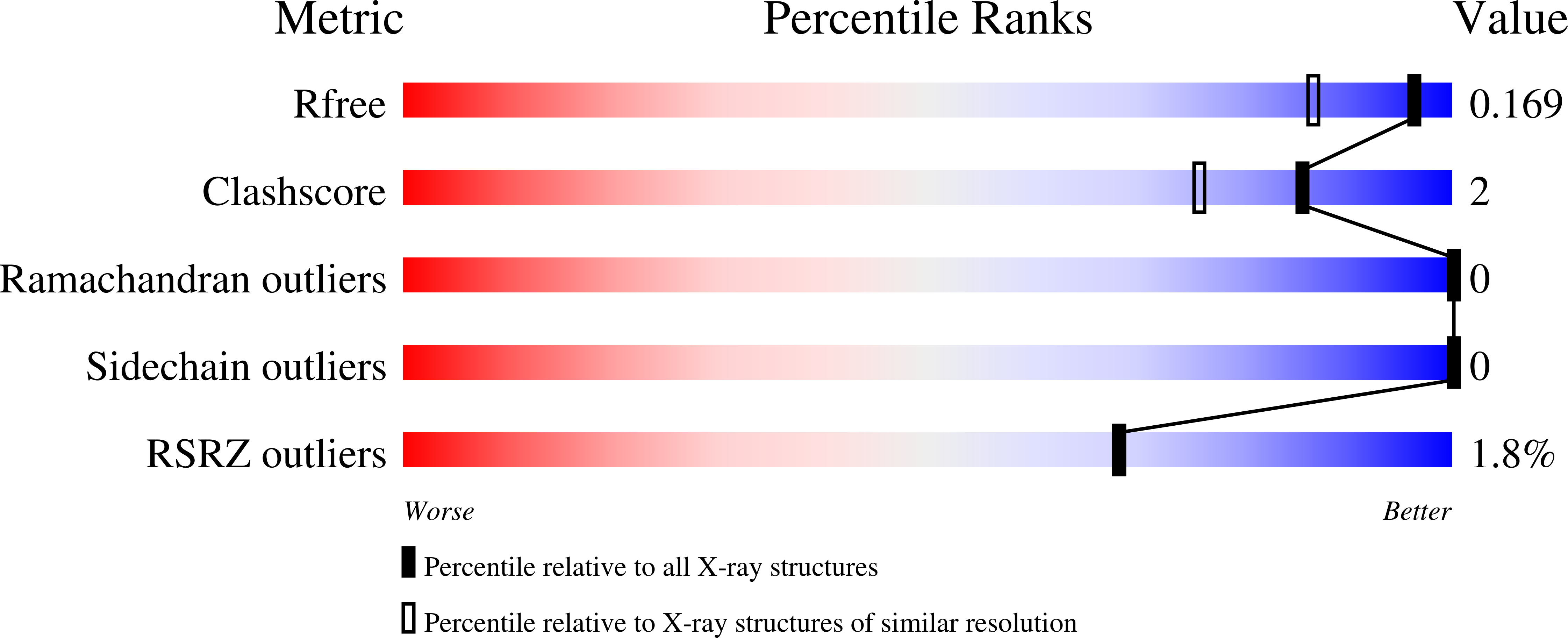
6O5W - PubMed Abstract:
Microrchidia 3 (MORC3), a human ATPase linked to several autoimmune disorders, has been characterized both as a negative and positive regulator of influenza A virus. Here, we report that the CW domain of MORC3 (MORC3-CW) is targeted by the C-terminal tail of the influenza H3N2 protein NS1. The crystal structure of the MORC3-CW:NS1 complex shows that NS1 occupies the same binding site in CW that is normally occupied by histone H3, a physiological ligand of MORC3-CW. Comparable binding affinities of MORC3-CW to H3 and NS1 peptides and to the adjacent catalytic ATPase domain suggest that the viral protein can compete with the host histone for the association with CW, releasing MORC3 autoinhibition and activating the catalytic function of MORC3. Our structural, biochemical, and cellular analyses suggest that MORC3 might affect the infectivity of influenza virus and therefore has a role in cell immune response.
Organizational Affiliation:
Department of Pharmacology, University of Colorado School of Medicine, Aurora, CO 80045, USA.