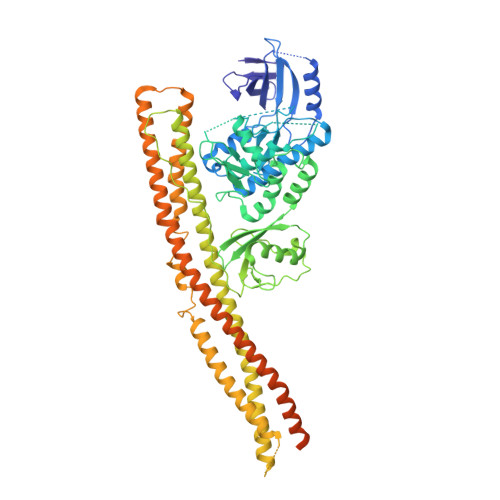
Structural basis of STING binding with and phosphorylation by TBK1.
Zhang, C., Shang, G., Gui, X., Zhang, X., Bai, X.C., Chen, Z.J.(2019) Nature 567: 394-398
- PubMed: 30842653
- DOI: https://doi.org/10.1038/s41586-019-1000-2
- Primary Citation of Related Structures:
6NT9 - PubMed Abstract:
The invasion of mammalian cytoplasm by microbial DNA from infectious pathogens or by self DNA from the nucleus or mitochondria represents a danger signal that alerts the host immune system 1 . Cyclic GMP-AMP synthase (cGAS) is a sensor of cytoplasmic DNA that activates the type-I interferon pathway 2 . On binding to DNA, cGAS is activated to catalyse the synthesis of cyclic GMP-AMP (cGAMP) from GTP and ATP 3 . cGAMP functions as a second messenger that binds to and activates stimulator of interferon genes (STING) 3-9 . STING then recruits and activates tank-binding kinase 1 (TBK1), which phosphorylates STING and the transcription factor IRF3 to induce type-I interferons and other cytokines 10,11 . However, how cGAMP-bound STING activates TBK1 and IRF3 is not understood. Here we present the cryo-electron microscopy structure of human TBK1 in complex with cGAMP-bound, full-length chicken STING. The structure reveals that the C-terminal tail of STING adopts a β-strand-like conformation and inserts into a groove between the kinase domain of one TBK1 subunit and the scaffold and dimerization domain of the second subunit in the TBK1 dimer. In this binding mode, the phosphorylation site Ser366 in the STING tail cannot reach the kinase-domain active site of bound TBK1, which suggests that STING phosphorylation by TBK1 requires the oligomerization of both proteins. Mutational analyses validate the interaction mode between TBK1 and STING and support a model in which high-order oligomerization of STING and TBK1, induced by cGAMP, leads to STING phosphorylation by TBK1.
Organizational Affiliation:
Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, TX, USA.