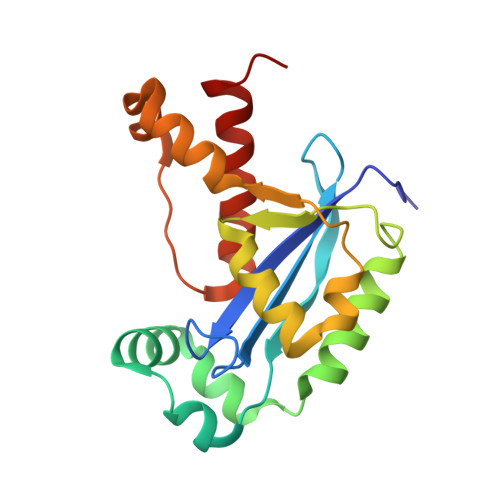
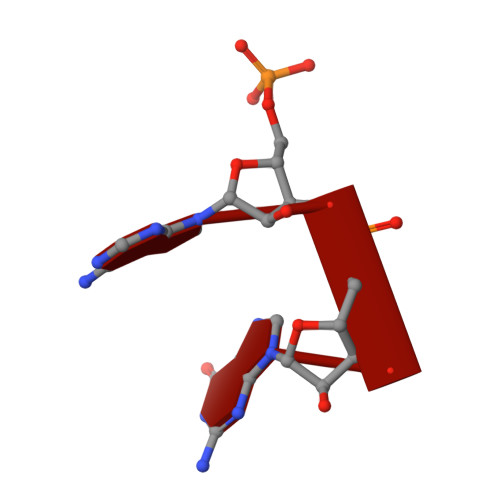
A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Kim, S.K., Lormand, J.D., Weiss, C.A., Eger, K.A., Turdiev, H., Turdiev, A., Winkler, W.C., Sondermann, H., Lee, V.T.(2019) Elife 8
- PubMed: 31225796
- DOI: https://doi.org/10.7554/eLife.46313
- Primary Citation of Related Structures:
6N6A, 6N6C, 6N6D, 6N6E, 6N6F, 6N6G, 6N6H, 6N6I, 6N6J, 6N6K - PubMed Abstract:
Degradation of RNA polymers, an ubiquitous process in all cells, is catalyzed by specific subsets of endo- and exoribonucleases that together recycle RNA fragments into nucleotide monophosphate. In γ-proteobacteria, 3-'5' exoribonucleases comprise up to eight distinct enzymes. Among them, Oligoribonuclease (Orn) is unique as its activity is required for clearing short RNA fragments, which is important for cellular fitness. However, the molecular basis of Orn's unique cellular function remained unclear. Here, we show that Orn exhibits exquisite substrate preference for diribonucleotides. Crystal structures of substrate-bound Orn reveal an active site optimized for diribonucleotides. While other cellular RNases process oligoribonucleotides down to diribonucleotide entities, Orn is the one and only diribonucleotidase that completes the terminal step of RNA degradation. Together, our studies indicate RNA degradation as a step-wise process with a dedicated enzyme for the clearance of a specific intermediate pool, diribonucleotides, that affects cellular physiology and viability.
Organizational Affiliation:
Department of Cell Biology and Molecular Genetics, University of Maryland, College Park, United States.