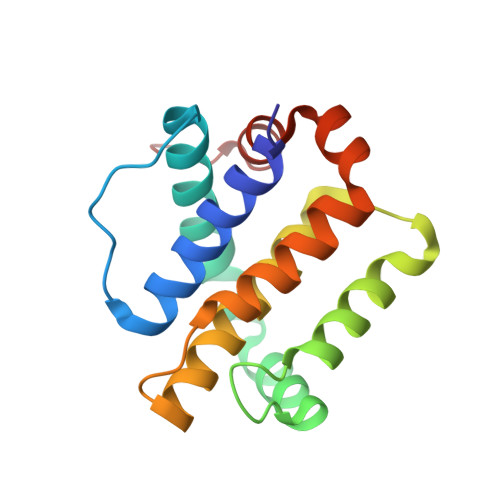
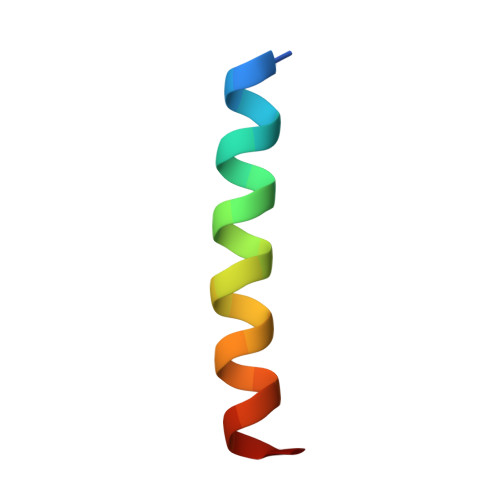
Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Frappier, V., Jenson, J.M., Zhou, J., Grigoryan, G., Keating, A.E.(2019) Structure 27: 606-617.e5
- PubMed: 30773399
- DOI: https://doi.org/10.1016/j.str.2019.01.008
- Primary Citation of Related Structures:
6MBB, 6MBC, 6MBD, 6MBE - PubMed Abstract:
Understanding the relationship between protein sequence and structure well enough to design new proteins with desired functions is a longstanding goal in protein science. Here, we show that recurring tertiary structural motifs (TERMs) in the PDB provide rich information for protein-peptide interaction prediction and design. TERM statistics can be used to predict peptide binding energies for Bcl-2 family proteins as accurately as widely used structure-based tools. Furthermore, design using TERM energies (dTERMen) rapidly and reliably generates high-affinity peptide binders of anti-apoptotic proteins Bfl-1 and Mcl-1 with just 15%-38% sequence identity to any known native Bcl-2 family protein ligand. High-resolution structures of four designed peptides bound to their targets provide opportunities to analyze the strengths and limitations of the computational design method. Our results support dTERMen as a powerful approach that can complement existing tools for protein engineering.
Organizational Affiliation:
Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.