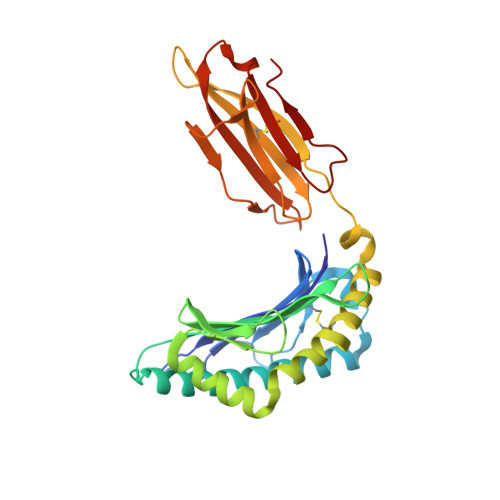
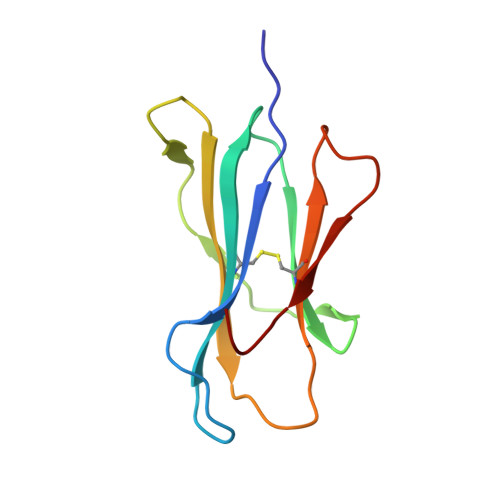
Crystal structures of lysophospholipid-bound MHC class I molecules.
Shima, Y., Morita, D., Mizutani, T., Mori, N., Mikami, B., Sugita, M.(2020) J Biol Chem 295: 6983-6991
- PubMed: 32269076
- DOI: https://doi.org/10.1074/jbc.RA119.011932
- Primary Citation of Related Structures:
6LAH, 6LAM, 6LB2, 6LT6 - PubMed Abstract:
Newly synthesized major histocompatibility complex (MHC) class I proteins are stabilized in the endoplasmic reticulum (ER) by binding 8-10-mer-long self-peptide antigens that are provided by transporter associated with antigen processing (TAP). These MHC class I:peptide complexes then exit the ER and reach the plasma membrane, serving to sustain the steady-state MHC class I expression on the cell surface. A novel subset of MHC class I molecules that preferentially bind lipid-containing ligands rather than conventional peptides was recently identified. The primate classical MHC class I allomorphs, Mamu-B*098 and Mamu-B*05104, are capable of binding the N -myristoylated 5-mer (C14-Gly-Gly-Ala-Ile-Ser) or 4-mer (C14-Gly-Gly-Ala-Ile) lipopeptides derived from the N -myristoylated SIV Nef protein, respectively, and of activating lipopeptide antigen-specific cytotoxic T lymphocytes. We herein demonstrate that Mamu-B*098 samples lysophosphatidylethanolamine and lysophosphatidylcholine containing up to a C20 fatty acid in the ER. The X-ray crystal structures of Mamu-B*098 and Mamu-B*05104 complexed with lysophospholipids at high resolution revealed that the B and D pockets in the antigen-binding grooves of these MHC class I molecules accommodate these lipids through a monoacylglycerol moiety. Consistent with the capacity to bind cellular lipid ligands, these two MHC class I molecules did not require TAP function for cell-surface expression. Collectively, these results indicate that peptide- and lipopeptide-presenting MHC class I subsets use distinct sources of endogenous ligands.
Organizational Affiliation:
Laboratory of Cell Regulation, Institute for Frontier Life and Medical Sciences, Kyoto University, 53 Kawahara-cho, Shogoin, Sakyo-ku, Kyoto 606-8507, Japan.