Structure of FliS in complex with flagellin and HP1076
Lam, W.W., Sun, K., Au, S.W.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Flagellar secretion chaperone FliS | 131 | Helicobacter pylori CPY1124 | Mutation(s): 0 Gene Names: fliS, HPCPY1124_0918 | 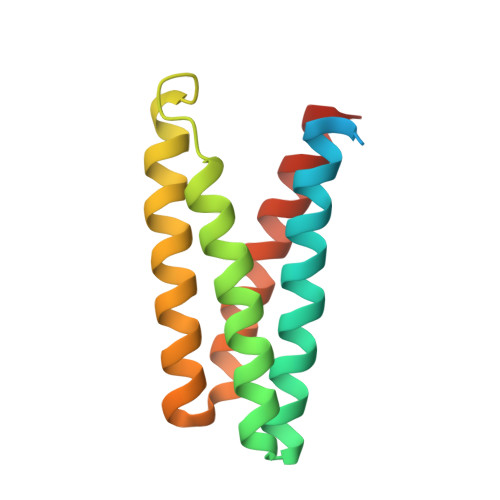 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Uncharacterized protein HP_1076 | 177 | Helicobacter pylori 26695 | Mutation(s): 0 Gene Names: HP_1076 | 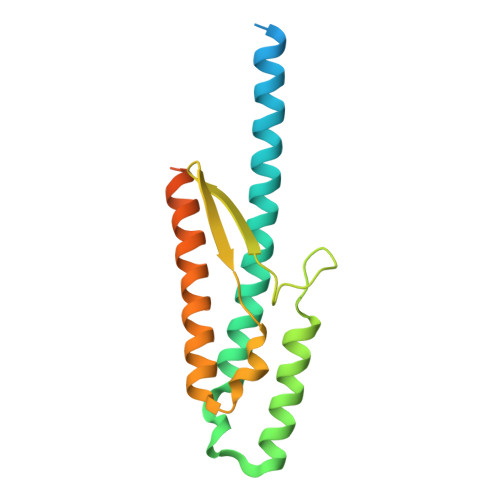 | |
UniProt | |||||
Find proteins for O25709 (Helicobacter pylori (strain ATCC 700392 / 26695)) Explore O25709 Go to UniProtKB: O25709 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O25709 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Flagellin | 105 | Helicobacter pylori | Mutation(s): 0 Gene Names: HPF63_0124 | 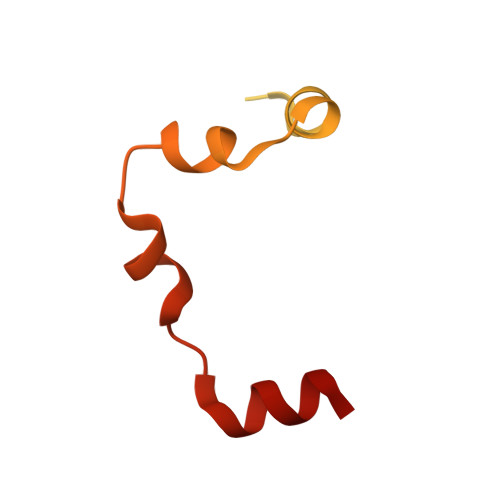 | |
UniProt | |||||
Find proteins for Q07911 (Helicobacter pylori (strain ATCC 700392 / 26695)) Explore Q07911 Go to UniProtKB: Q07911 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q07911 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 103.33 | α = 90 |
| b = 103.33 | β = 90 |
| c = 144.211 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| MOSFLM | data reduction |
| SCALA | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| The University Grants Committee, Research Grants Council (RGC) | Hong Kong | 14100114 |