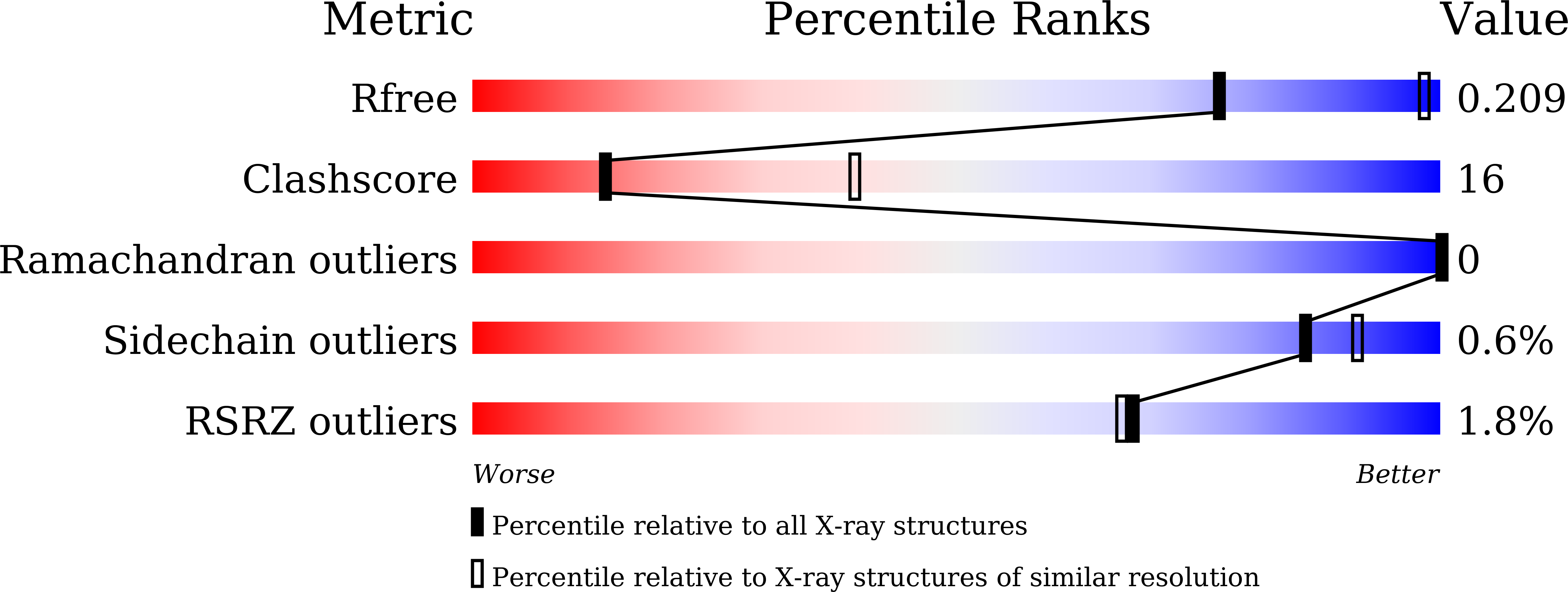
Structural and Biophysical Analyses of Human N-Myc Downstream-Regulated Gene 3 (NDRG3) Protein.
Kim, K.R., Kim, K.A., Park, J.S., Jang, J.Y., Choi, Y., Lee, H.H., Lee, D.C., Park, K.C., Yeom, Y.I., Kim, H.J., Han, B.W.(2020) Biomolecules 10
- PubMed: 31935861
- DOI: https://doi.org/10.3390/biom10010090
- Primary Citation of Related Structures:
6L4B, 6L4G, 6L4H - PubMed Abstract:
The N-Myc downstream-regulated gene (NDRG) family belongs to the α/β-hydrolase fold and is known to exert various physiologic functions in cell proliferation, differentiation, and hypoxia-induced cancer metabolism. In particular, NDRG3 is closely related to proliferation and migration of prostate cancer cells, and recent studies reported its implication in lactate-triggered hypoxia responses or tumorigenesis. However, the underlying mechanism for the functions of NDRG3 remains unclear. Here, we report the crystal structure of human NDRG3 at 2.2 Å resolution, with six molecules in an asymmetric unit. While NDRG3 adopts the α/β-hydrolase fold, complete substitution of the canonical catalytic triad residues to non-reactive residues and steric hindrance around the pseudo-active site seem to disable the α/β-hydrolase activity. While NDRG3 shares a high similarity to NDRG2 in terms of amino acid sequence and structure, NDRG3 exhibited remarkable structural differences in a flexible loop corresponding to helix α6 of NDRG2 that is responsible for tumor suppression. Thus, this flexible loop region seems to play a distinct role in oncogenic progression induced by NDRG3. Collectively, our studies could provide structural and biophysical insights into the molecular characteristics of NDRG3.
Organizational Affiliation:
Research Institute of Pharmaceutical Sciences, College of Pharmacy, Seoul National University, Seoul 08826, Korea.