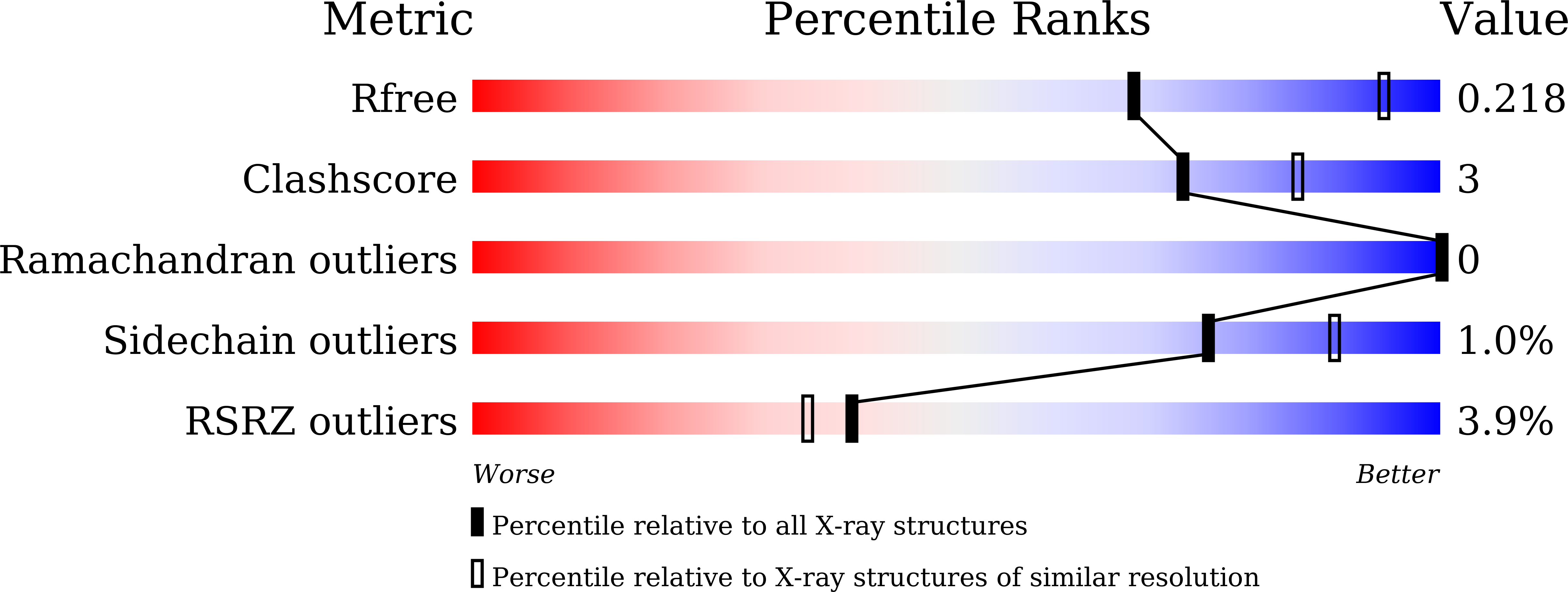
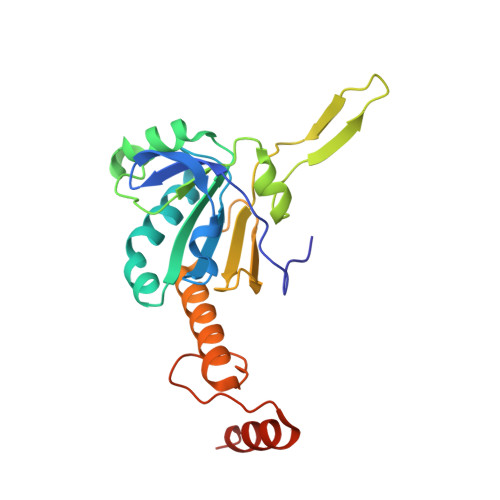
Crystal structure of Akkermansia muciniphila peroxiredoxin reveals a novel regulatory mechanism of typical 2-Cys Prxs by a distinct loop.
Li, M., Wang, J., Xu, W., Wang, Y., Zhang, M., Wang, M.(2020) FEBS Lett 594: 1550-1563
- PubMed: 32027024
- DOI: https://doi.org/10.1002/1873-3468.13753
- Primary Citation of Related Structures:
6KHX - PubMed Abstract:
Peroxiredoxins (Prxs) are thiol-specific antioxidant proteins commonly found in organisms that protect cells from the damage of reactive oxygen species produced by metabolism and that participate in cell signaling. The Prx from the bacterium Akkermansia muciniphila (AmPrx) is a typical 2-Cys Prx characterized by two conserved cysteines: Cys49 and Cys183. Here, we verified the peroxidase activity of AmPrx and determined its crystal structure in reduced form, which is a doughnut-shaped decamer composed of five dimers. Particularly, a distinct loop between the α4 helix and β6 strand is involved in the decameric interaction. Deletion of this loop destroys the decameric structure and significantly decreases the peroxidase activity of AmPrx. Our findings reveal a novel regulatory mechanism of typical 2-Cys Prx, in which the α4-β6 loop affects the assembly of Prx and, therefore, regulates its peroxidase activity.
Organizational Affiliation:
School of Life Sciences, Anhui University, Hefei, China.