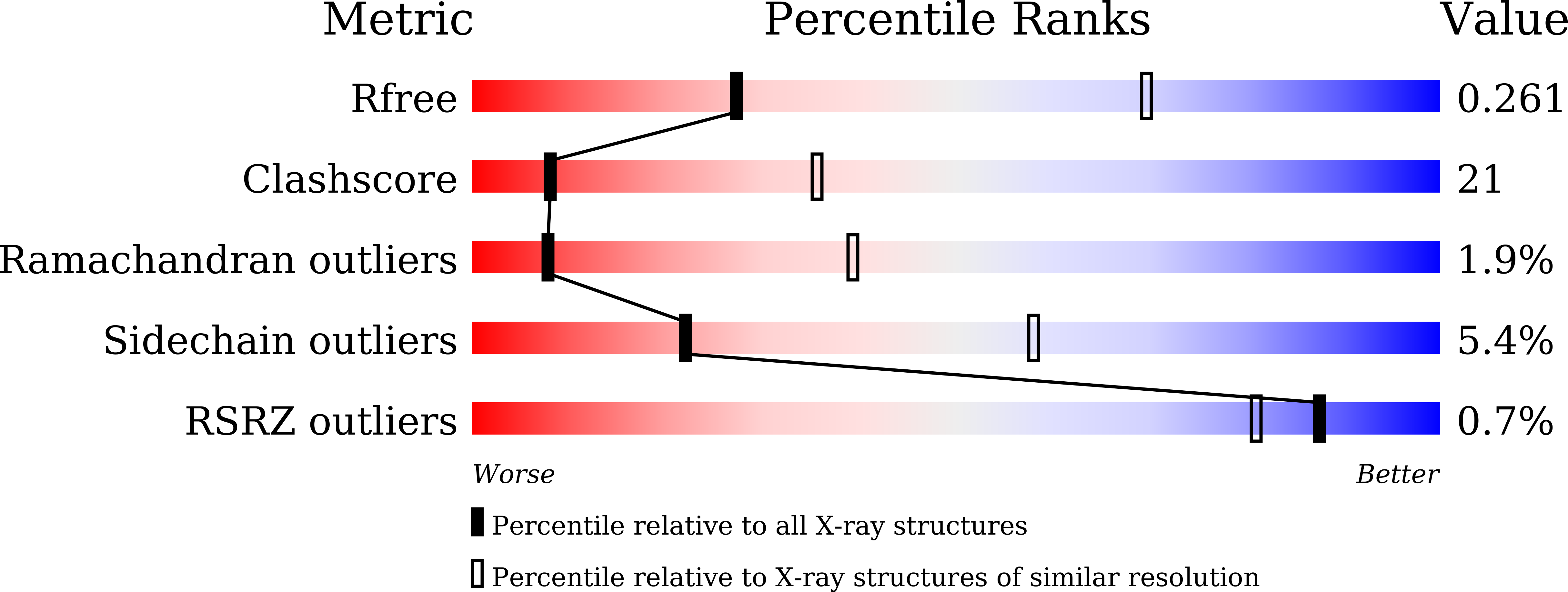
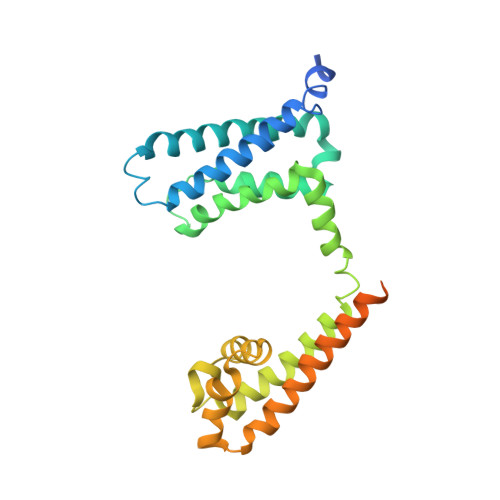
Structural Basis for Diltiazem Block of a Voltage-Gated Ca2+Channel.
Tang, L., Gamal El-Din, T.M., Lenaeus, M.J., Zheng, N., Catterall, W.A.(2019) Mol Pharmacol 96: 485-492
- PubMed: 31391290
- DOI: https://doi.org/10.1124/mol.119.117531
- Primary Citation of Related Structures:
6KE5, 6KEB - PubMed Abstract:
Diltiazem is a widely prescribed Ca 2+ antagonist drug for cardiac arrhythmia, hypertension, and angina pectoris. Using the ancestral Ca V channel construct Ca V Ab as a molecular model for X-ray crystallographic analysis, we show here that diltiazem targets the central cavity of the voltage-gated Ca 2+ channel underneath its selectivity filter and physically blocks ion conduction. The diltiazem-binding site overlaps with the receptor site for phenylalkylamine Ca 2+ antagonist drugs such as verapamil. The dihydropyridine Ca 2+ channel blocker amlodipine binds at a distinct site and allosterically modulates the binding sites for diltiazem and Ca 2+ Our studies resolve two distinct binding poses for diltiazem in the absence and presence of amlodipine. The binding pose in the presence of amlodipine may mimic a high-affinity binding configuration induced by voltage-dependent inactivation, which is favored by dihydropyridine binding. In this binding pose, the tertiary amino group of diltiazem projects upward into the inner end of the ion selectivity filter, interacts with ion coordination Site 3 formed by the backbone carbonyls of T175, and alters binding of Ca 2+ to ion coordination Sites 1 and 2. Altogether, our results define the receptor site for diltiazem and elucidate the mechanisms for pore block and allosteric modulation by other Ca 2+ channel-blocking drugs at the atomic level. SIGNIFICANCE STATEMENT: Calcium antagonist drugs that block voltage-gated calcium channels in heart and vascular smooth muscle are widely used in the treatment of cardiovascular diseases. Our results reveal the chemical details of diltiazem binding in a blocking position in the pore of a model calcium channel and show that binding of another calcium antagonist drug alters binding of diltiazem and calcium. This structural information defines the mechanism of drug action at the atomic level and provides a molecular template for future drug discovery.
Organizational Affiliation:
Department of Neurology, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University and Collaborative Innovation Center for Biotherapy, Chengdu, Sichuan, China (L.T.); and Department of Pharmacology (L.T., T.M.G.E.-D., M.J.L., N.Z., W.A.C.), Division of General Internal Medicine, Department of Medicine (M.J.L.), and Howard Hughes Medical Institute (N.Z.), University of Washington, Seattle, Washington ltang@scu.edu.cn.