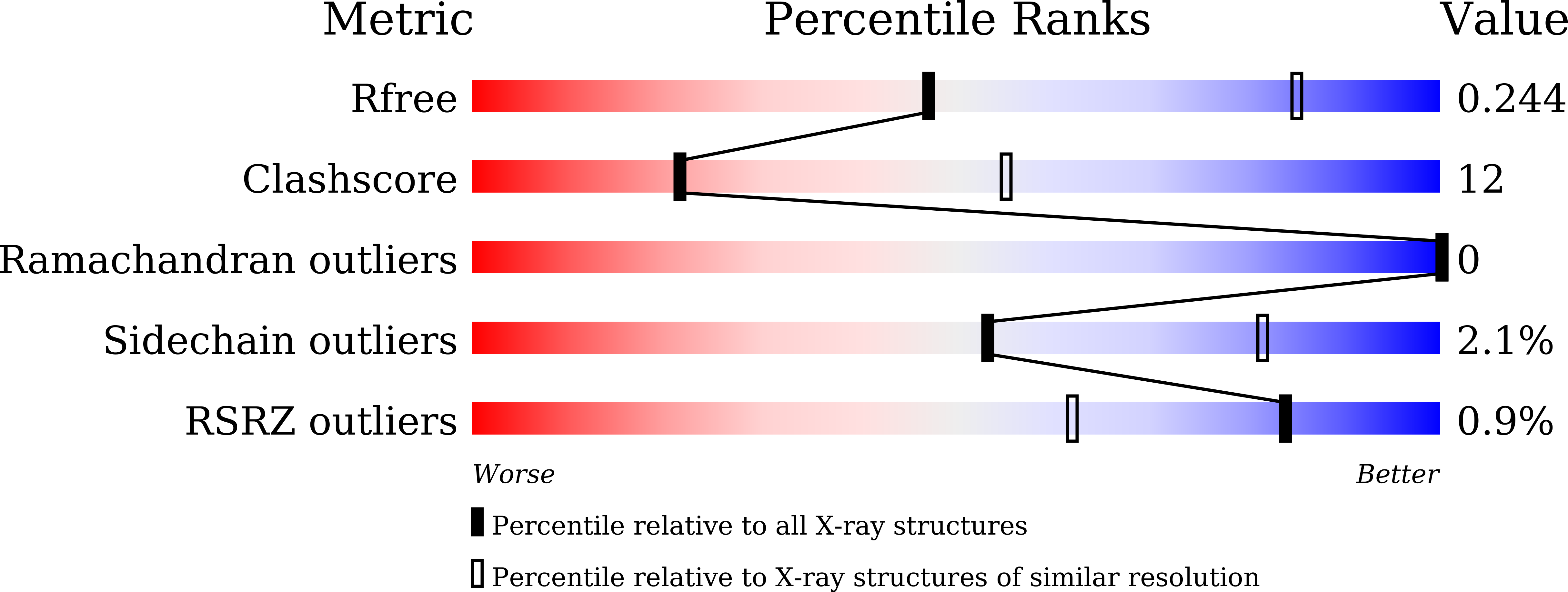
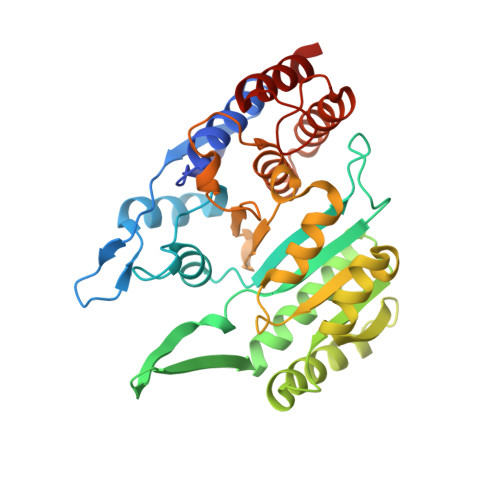
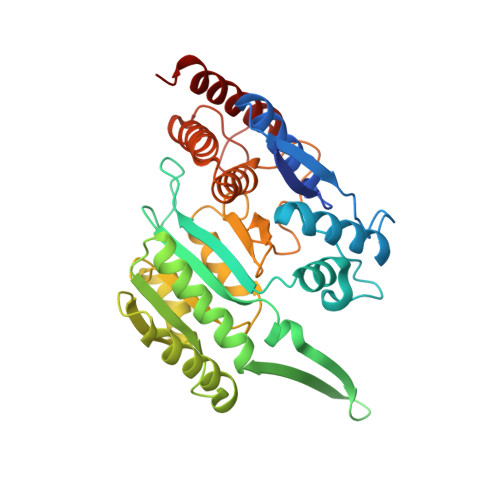
Molecular basis for the function of the alpha beta heterodimer of human NAD-dependent isocitrate dehydrogenase.
Sun, P., Ma, T., Zhang, T., Zhu, H., Zhang, J., Liu, Y., Ding, J.(2019) J Biol Chem 294: 16214-16227
- PubMed: 31515270
- DOI: https://doi.org/10.1074/jbc.RA119.010099
- Primary Citation of Related Structures:
6KDE, 6KDF, 6KDY, 6KE3 - PubMed Abstract:
Mammalian mitochondrial NAD-dependent isocitrate dehydrogenase (NAD-IDH) catalyzes the decarboxylation of isocitrate into α-ketoglutarate in the tricarboxylic acid cycle. It exists as the α 2 βγ heterotetramer composed of the αβ and αγ heterodimers. Different from the αγ heterodimer that can be allosterically activated by CIT and ADP, the αβ heterodimer cannot be allosterically regulated by the activators; however, the molecular mechanism is unclear. We report here the crystal structures of the αβ heterodimer of human NAD-IDH with the α subunit in apo form and in Ca 2+ -bound, NAD-bound, and NADH-bound forms. Structural analyses and comparisons reveal that the αβ heterodimer has a similar yet more compact overall structure compared with the αγ heterodimer and contains a pseudo-allosteric site that is structurally different from the allosteric site. In particular, the β3-α3 and β12-α8 loops of the β subunit at the pseudo-allosteric site adopt significantly different conformations from those of the γ subunit at the allosteric site and hence impede the binding of the activators, explaining why the αβ heterodimer cannot be allosterically regulated by the activators. The structural data also show that NADH can compete with NAD to bind to the active site and inhibits the activity of the αβ heterodimer. These findings together with the biochemical data reveal the molecular basis for the function of the αβ heterodimer of human NAD-IDH.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Institute of Biochemistry and Cell Biology, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yue-Yang Road, Shanghai 200031, China.