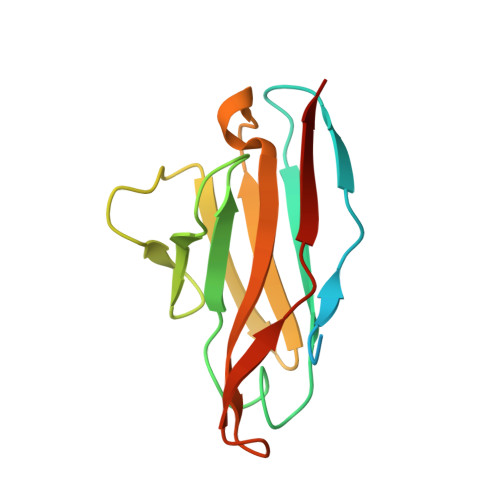
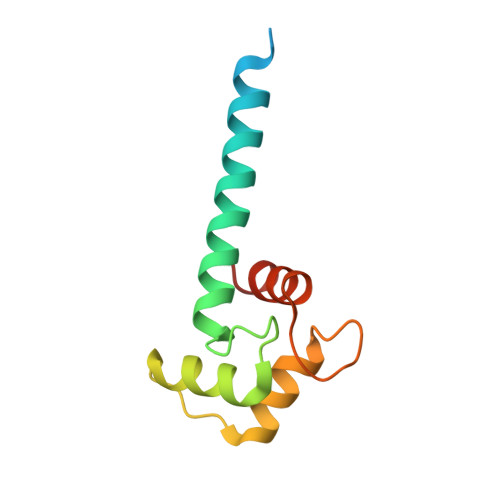
Application of antihelix antibodies in protein structure determination.
Kim, J.W., Kim, S., Lee, H., Cho, G., Kim, S.C., Lee, H., Jin, M.S., Lee, J.O.(2019) Proc Natl Acad Sci U S A 116: 17786-17791
- PubMed: 31371498
- DOI: https://doi.org/10.1073/pnas.1910080116
- Primary Citation of Related Structures:
6K3M, 6K64, 6K65, 6K67, 6K68, 6K69, 6K6A, 6K6B - PubMed Abstract:
Antibodies are indispensable tools in protein engineering and structural biology. Antibodies suitable for structural studies should recognize the 3-dimensional (3D) conformations of target proteins. Generating such antibodies and characterizing their complexes with antigens take a significant amount of time and effort. Here, we show that we can expand the application of well-characterized antibodies by "transplanting" the epitopes that they recognize to proteins with completely different structures and sequences. Previously, several antibodies have been shown to recognize the alpha-helical conformation of antigenic peptides. We demonstrate that these antibodies can be made to bind to a variety of unrelated "off-target" proteins by modifying amino acids in the preexisting alpha helices of such proteins. Using X-ray crystallography, we determined the structures of the engineered protein-antibody complexes. All of the antibodies bound to the epitope-transplanted proteins, forming accurately predictable structures. Furthermore, we showed that binding of these antihelix antibodies to the engineered target proteins can modulate their catalytic activities by trapping them in selected functional states. Our method is simple and efficient, and it will have applications in protein X-ray crystallography, electron microscopy, and nanotechnology.
Organizational Affiliation:
Department of Life Sciences, Pohang University of Science and Technology, Nam-gu, Pohang 37673, Korea.