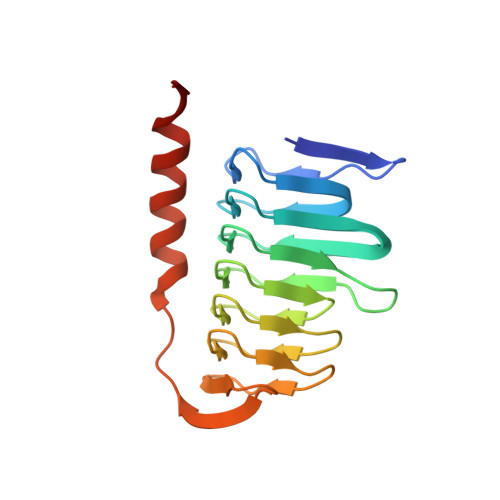
Molecular structure of thermostable and zinc-ion-binding gamma-class carbonic anhydrases.
Wang, W., Zhang, Y., Wang, L., Jing, Q., Wang, X., Xi, X., Zhao, X., Wang, H.(2019) Biometals 32: 317-328
- PubMed: 30895492
- DOI: https://doi.org/10.1007/s10534-019-00190-8
- Primary Citation of Related Structures:
6IVE - PubMed Abstract:
The γ-class carbonic anhydrases (γ-CAs) mainly come from methanogens methane-producing bacteria that grow in hot springs and catalyze the interconversion of carbon dioxide and water to bicarbonate and protons. Here, the γ-CA from Thermus thermophilus HB8 (γ-TtCA) was expressed and purified, its crystal structure was determined at 2.3 Å resolution in space group P1. The asymmetric unit contains two trimers and six catalytic Zn 2+ . In general, the fold of the protein is similar to those of homologous enzymes from Geobacillus Kaustophilus, Bacillus Cereus, Methanosarcina Thermophila and others. Each monomer comprises a triangular prism-like structure consisting of a left-handed β-helix and a C-terminal α-helix. The catalytic Zn 2+ bound to three histidines and a phosphate radical in a tetrahedral fashion. It is located at the interface between the two monomers. Inductively coupled plasma mass spectrometry measurements further suggest that the molar ratio of zinc ions and protein molecules is 1:1. The structure revealed a novel different region situated between the left-handed β-helix and the C-terminal α-helix. Compared to previously reported structures, half of the C-terminal α-helix was replaced with a long loop in this structure. The purified γ-TtCA exhibits no significant carbonic anhydrase activity compared to α-class carbonic anhydrases. This study provides insight into the structural diversity of γ-CAs with potential function for γ-CAs.
Organizational Affiliation:
Key Laboratory of Chemical Biology and Molecular Engineering of Education Ministry, Institute of Molecular Science, Shanxi University, Taiyuan, 030006, China. wangwm@sxu.edu.cn.