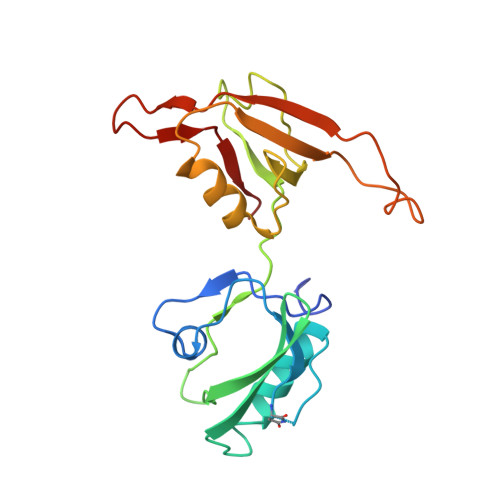
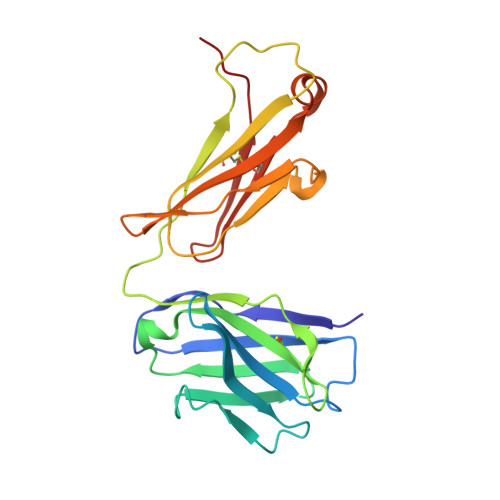
Application of the NZ-1 Fab as a crystallization chaperone for PA tag-inserted target proteins.
Tamura, R., Oi, R., Akashi, S., Kaneko, M.K., Kato, Y., Nogi, T.(2019) Protein Sci 28: 823-836
- PubMed: 30666745
- DOI: https://doi.org/10.1002/pro.3580
- Primary Citation of Related Structures:
6AKQ, 6AL0, 6AL1, 6ICC, 6ICF - PubMed Abstract:
An antibody fragment that recognizes the tertiary structure of a target protein with high affinity can be utilized as a crystallization chaperone. Difficulties in establishing conformation-specific antibodies, however, limit the applicability of antibody fragment-assisted crystallization. Here, we attempted to establish an alternative method to promote the crystallization of target proteins using an already established anti-tag antibody. The monoclonal antibody NZ-1 recognizes the PA tag with an extremely high affinity. It was also established that the PA tag is accommodated in the antigen-binding pocket in a bent conformation, compatible with an insertion into loop regions on the target. We, therefore, explored the application of NZ-1 Fab as a crystallization chaperone that complexes with a target protein displaying a PA tag. Specifically, we inserted the PA tag into the β-hairpins of the PDZ tandem fragment of a bacterial Site-2 protease. We crystallized the PA-inserted PDZ tandem mutants with the NZ-1 Fab and solved the co-crystal structure to analyze their interaction modes. Although the initial insertion designs produced only moderate-resolution structures, eliminating the solvent-accessible space between the NZ-1 Fab and target PDZ tandem improved the diffraction qualities remarkably. Our results demonstrate that the NZ-1-PA system efficiently promotes crystallization of the target protein. The present work also suggests that β-hairpins are suitable sites for the PA insertion because the PA tag contains a Pro-Gly sequence with a propensity for a β-turn conformation.
Organizational Affiliation:
Graduate School of Medical Life Science, Yokohama City University, Yokohama, Japan.