Optimization of Potent and Selective Ataxia Telangiectasia-Mutated Inhibitors Suitable for a Proof-of-Concept Study in Huntington's Disease Models.
Toledo-Sherman, L., Breccia, P., Cachope, R., Bate, J.R., Angulo-Herrera, I., Wishart, G., Matthews, K.L., Martin, S.L., Cox, H.C., McAllister, G., Penrose, S.D., Vater, H., Esmieu, W., Van de Poel, A., Van de Bospoort, R., Strijbosch, A., Lamers, M., Leonard, P., Jarvis, R.E., Blackaby, W., Barnes, K., Eznarriaga, M., Dowler, S., Smith, G.D., Fischer, D.F., Lazari, O., Yates, D., Rose, M., Jang, S.W., Munoz-Sanjuan, I., Dominguez, C.(2019) J Med Chem 62: 2988-3008
- PubMed: 30840447
- DOI: https://doi.org/10.1021/acs.jmedchem.8b01819
- Primary Citation of Related Structures:
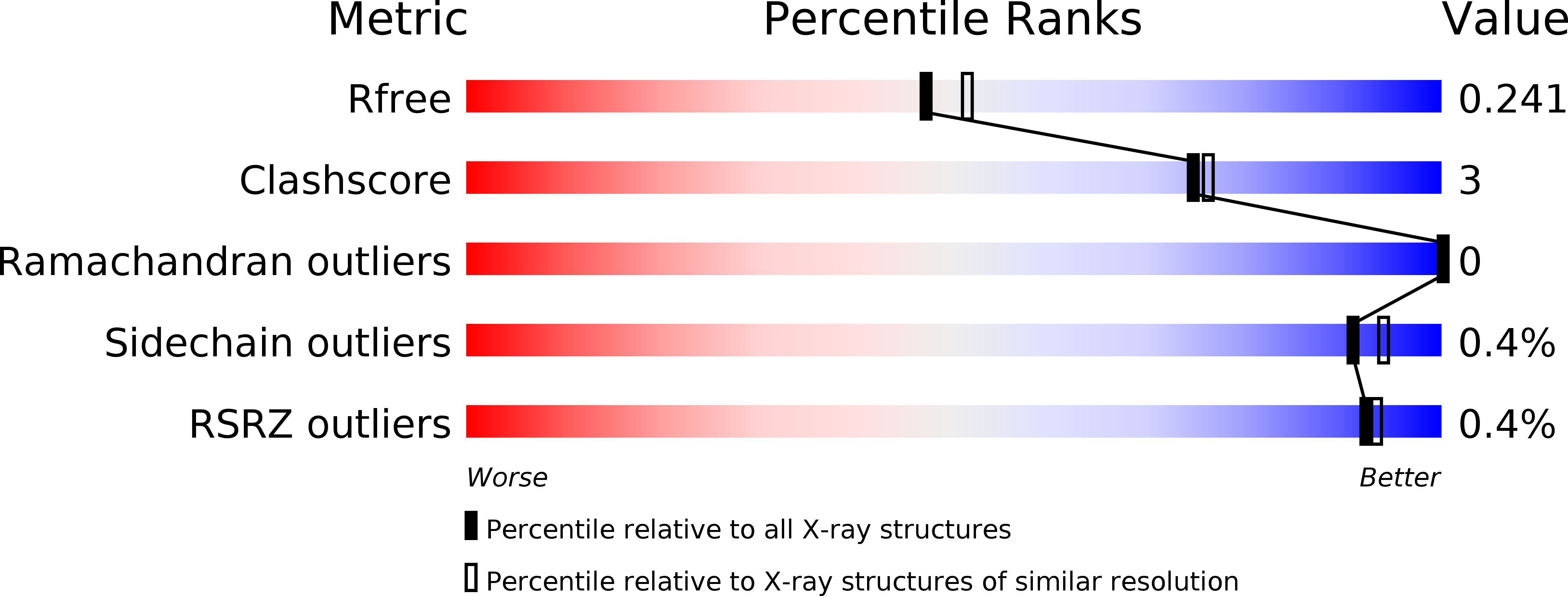
6I3U - PubMed Abstract:
Genetic and pharmacological evidence indicates that the reduction of ataxia telangiectasia-mutated (ATM) kinase activity can ameliorate mutant huntingtin (mHTT) toxicity in cellular and animal models of Huntington's disease (HD), suggesting that selective inhibition of ATM could provide a novel clinical intervention to treat HD. Here, we describe the development and characterization of ATM inhibitor molecules to enable in vivo proof-of-concept studies in HD animal models. Starting from previously reported ATM inhibitors, we aimed with few modifications to increase brain exposure by decreasing P-glycoprotein liability while maintaining potency and selectivity. Here, we report brain-penetrant ATM inhibitors that have robust pharmacodynamic (PD) effects consistent with ATM kinase inhibition in the mouse brain and an understandable pharmacokinetic/PD (PK/PD) relationship. Compound 17 engages ATM kinase and shows robust dose-dependent inhibition of X-ray irradiation-induced KAP1 phosphorylation in the mouse brain. Furthermore, compound 17 protects against mHTT (Q73)-induced cytotoxicity in a cortical-striatal cell model of HD.
Organizational Affiliation:
CHDI Management/CHDI Foundation , 6080 Center Drive , Los Angeles , California 90045 , United States.