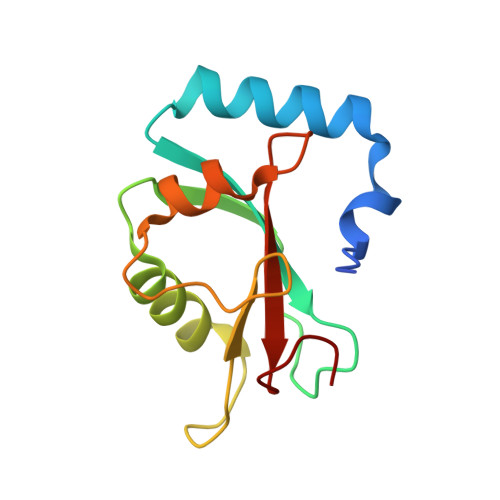
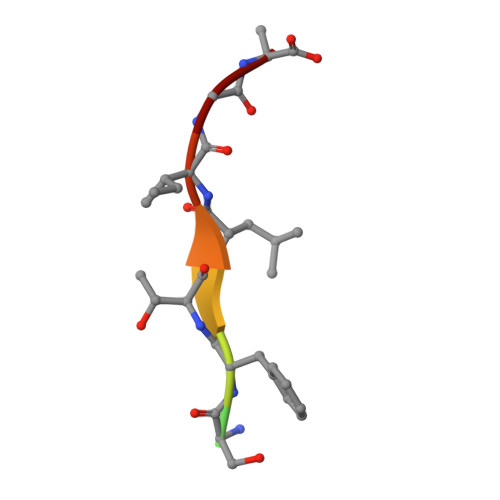
Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Birgisdottir, A.B., Mouilleron, S., Bhujabal, Z., Wirth, M., Sjottem, E., Evjen, G., Zhang, W., Lee, R., O'Reilly, N., Tooze, S.A., Lamark, T., Johansen, T.(2019) Autophagy 15: 1333-1355
- PubMed: 30767700
- DOI: https://doi.org/10.1080/15548627.2019.1581009
- Primary Citation of Related Structures:
6HOG, 6HOH, 6HOI, 6HOJ, 6HOK, 6HOL - PubMed Abstract:
Autophagosome formation depends on a carefully orchestrated interplay between membrane-associated protein complexes. Initiation of macroautophagy/autophagy is mediated by the ULK1 (unc-51 like autophagy activating kinase 1) protein kinase complex and the autophagy-specific class III phosphatidylinositol 3-kinase complex I (PtdIns3K-C1). The latter contains PIK3C3/VPS34, PIK3R4/VPS15, BECN1/Beclin 1 and ATG14 and phosphorylates phosphatidylinositol to generate phosphatidylinositol 3-phosphate (PtdIns3P). Here, we show that PIK3C3, BECN1 and ATG14 contain functional LIR motifs and interact with the Atg8-family proteins with a preference for GABARAP and GABARAPL1. High resolution crystal structures of the functional LIR motifs of these core components of PtdIns3K-C1were obtained. Variation in hydrophobic pocket 2 (HP2) may explain the specificity for the GABARAP family. Mutation of the LIR motif in ATG14 did not prevent formation of the PtdIns3K-C1 complex, but blocked colocalization with MAP1LC3B/LC3B and impaired mitophagy. The ULK-mediated phosphorylation of S29 in ATG14 was strongly dependent on a functional LIR motif in ATG14. GABARAP-preferring LIR motifs in PIK3C3, BECN1 and ATG14 may, via coincidence detection, contribute to scaffolding of PtdIns3K-C1 on membranes for efficient autophagosome formation. Abbreviations: ATG: autophagy-related; BafA1: bafilomycin A 1 ; GABARAP: GABA type A receptor-associated protein; GABARAPL1: GABA type A receptor associated protein like 1; GFP: enhanced green fluorescent protein; KO: knockout; LDS: LIR docking site; LIR: LC3-interacting region; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; PIK3C3: phosphatidylinositol 3-kinase catalytic subunit type 3; PIK3R4: phosphoinositide-3-kinase regulatory subunit 4; PtdIns3K: phosphatidylinositol 3-kinase; PtdIns3P: phosphatidylinositol-3-phosphate; SQSTM1/p62: sequestosome 1; VPS: Vacuolar protein sorting; ULK: unc-51 like autophagy activating kinase.
Organizational Affiliation:
a Molecular Cancer Research Group, Department of Medical Biology , University of Tromsø -The Arctic University of Norway , Tromsø , Norway.