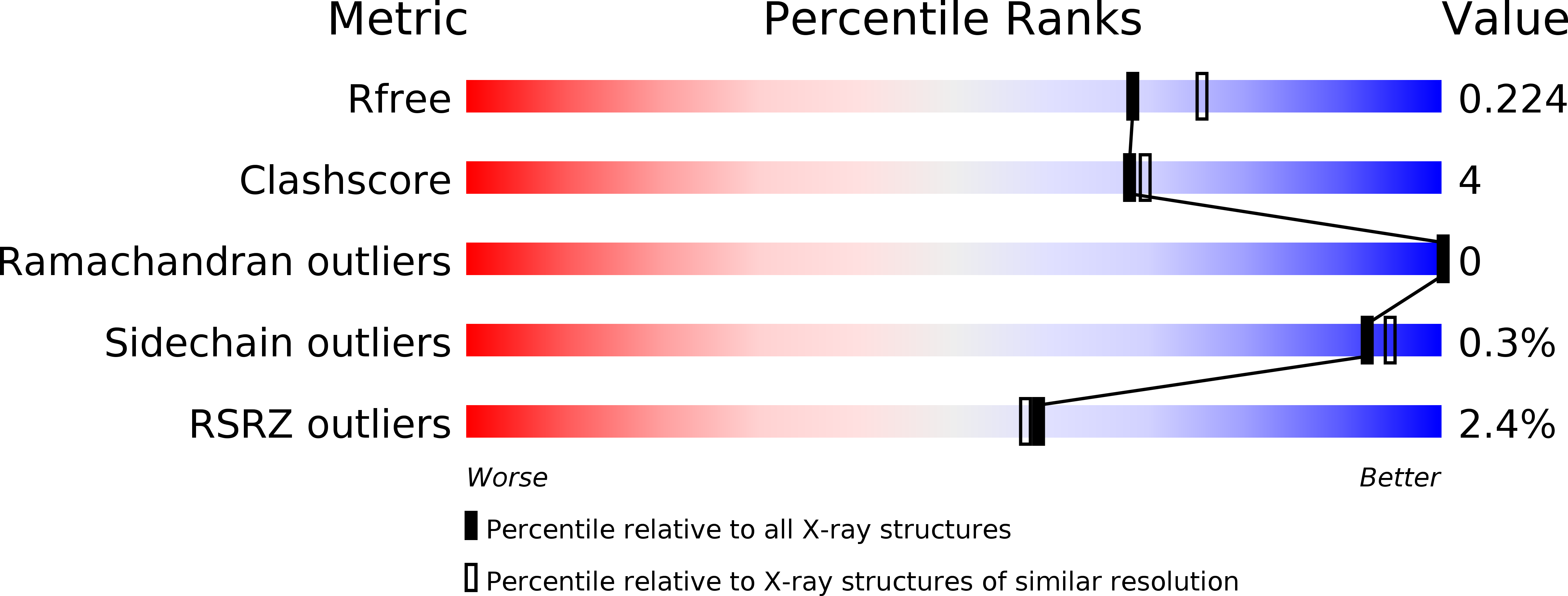
Inhibitory Properties of ATP-Competitive Coumestrol and Boldine Are Correlated to Different Modulations of CK2 Flexibility.
Battistutta, R., Lolli, G.(2019) J Nat Prod 82: 1014-1018
- PubMed: 30840451
- DOI: https://doi.org/10.1021/acs.jnatprod.8b00889
- Primary Citation of Related Structures:
6HNW, 6HNY - PubMed Abstract:
Casein kinase 2 (CK2) is an anti-apoptotic cancer-sustaining protein kinase. Its crystallographic structures with the natural compounds coumestrol, a phytoestrogen, and boldine, an alkaloid, are reported. Coumestrol shows different inhibitory activity against the isolated catalytic α-subunit and the α 2 β 2 holoenzyme and is able to discriminate between two conformations of the hinge/αD region, whose intrinsic flexibility is a relevant selectivity determinant among kinases. Boldine explores a small cavity at the bottom of the ATP-binding pocket through a local deviation from planarity, a unique case among CK2 inhibitors. The two compounds have different impacts on protein flexibility, which correlate with their different properties.
Organizational Affiliation:
Department of Chemical Sciences , University of Padua and Institute of Biomolecular Chemistry, National Research Council (CNR) , 35131 Padua , Italy.