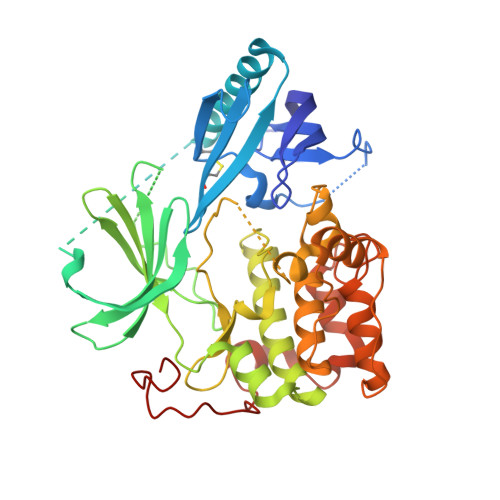
Structural and chemical insights into the covalent-allosteric inhibition of the protein kinase Akt.
Uhlenbrock, N., Smith, S., Weisner, J., Landel, I., Lindemann, M., Le, T.A., Hardick, J., Gontla, R., Scheinpflug, R., Czodrowski, P., Janning, P., Depta, L., Quambusch, L., Muller, M.P., Engels, B., Rauh, D.(2019) Chem Sci 10: 3573-3585
- PubMed: 30996949
- DOI: https://doi.org/10.1039/c8sc05212c
- Primary Citation of Related Structures:
6HHG, 6HHH, 6HHI, 6HHJ - PubMed Abstract:
The Ser/Thr kinase Akt (Protein Kinase B/PKB) is a master switch in cellular signal transduction pathways. Its downstream signaling influences cell proliferation, cell growth, and apoptosis, rendering Akt a prominent drug target. The unique activation mechanism of Akt involves a change of the relative orientation of its N-terminal pleckstrin homology (PH) and the kinase domain and makes this kinase suitable for highly specific allosteric modulation. Here we present a unique set of crystal structures of covalent-allosteric interdomain inhibitors in complex with full-length Akt and report the structure-based design, synthesis, biological and pharmacological evaluation of a focused library of these innovative inhibitors.
Organizational Affiliation:
Faculty of Chemistry and Chemical Biology , TU Dortmund University , Drug Discovery Hub Dortmund (DDHD) am Zentrum für integrierte Wirkstoffforschung (ZIW) , Otto-Hahn-Strasse 4a , 44227 Dortmund , Germany . Email: daniel.rauh@tu-dortmund.de ; http://www.ddhdortmund.de ; www.twitter.com/DDHDortmund.