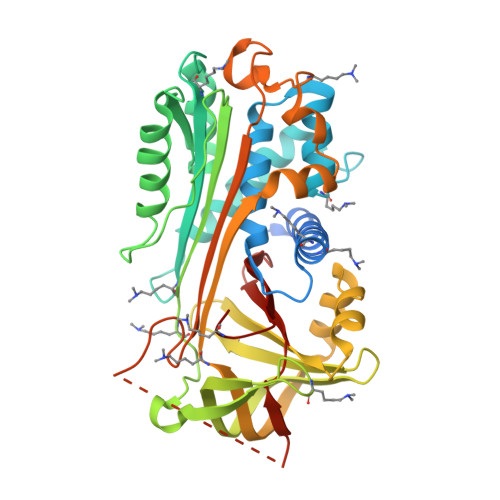
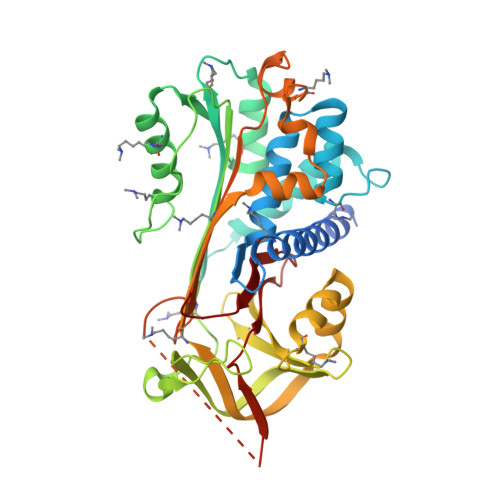
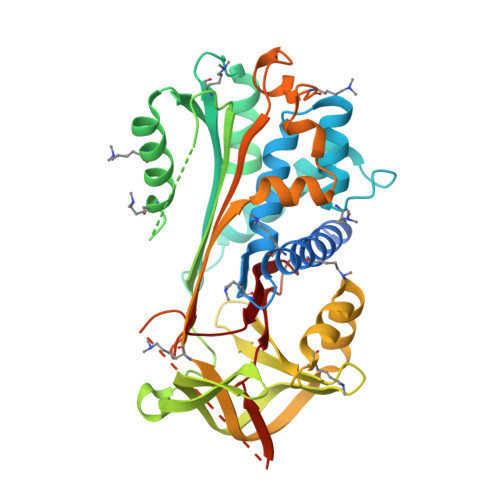
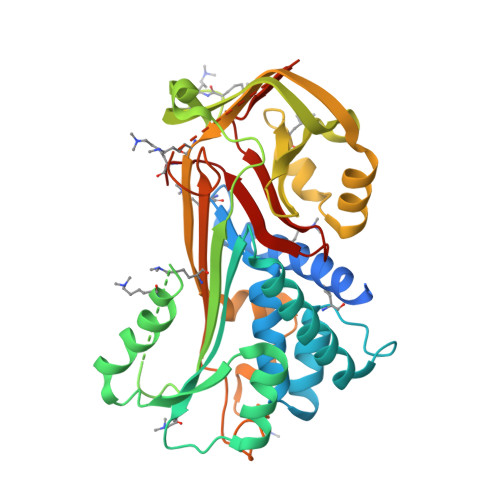
NewBG: A surrogate corticosteroid-binding globulin with an unprecedentedly high ligand release efficacy.
Gardill, B.R., Schmidt, K., Muller, Y.A.(2019) J Struct Biol 207: 169-182
- PubMed: 31103428
- DOI: https://doi.org/10.1016/j.jsb.2019.05.006
- Primary Citation of Related Structures:
6HGD, 6HGE, 6HGF, 6HGG, 6HGH, 6HGI, 6HGJ, 6HGK, 6HGL, 6HGM, 6HGN - PubMed Abstract:
The introduction of ligand-binding sites into proteins and the engineering of molecular allosteric coupling pathways are topical issues in protein design. Here, we show that these issues can be addressed concurrently, using the serpin human α1-antichymotrypsin (ACT) as a model. We have introduced up to 15 amino acid substitutions into ACT, converting it into a surrogate corticosteroid-binding globulin (CBG), thereby creating a new binding globulin (NewBG). Human CBG and ACT share 46% sequence identity, and CBG served as the blue-print for our design, which was guided by side-chain-packing calculations, ITC measurements and crystal structure determinations. Upon transfer of ligand-interacting residues from CBG to ACT and mutation of specific second shell residues, a NewBG variant was obtained, which binds cortisol with 1.5 µM affinity. This novel serpin (NewBG-III) binds cortisol with a 33-fold lower affinity than CBG, but shares a similar ligand-binding profile and binding mode when probed with different steroid ligands and site-directed mutagenesis. An additional substitution, i.e. A349R, created NewBG-III-allo, which introduced an allosteric coupling between ligand binding and the serpin-like S-to-R transition in ACT. In NewBG-III-allo, the proteinase-triggered S-to-R transition leads to a greater than 200-fold reduction in ligand affinity, and crystal structures suggest that this is mediated by the L55V and A349R substitutions. This reduction significantly exceeds the 10-fold reduction in binding affinity observed in human CBG.
Organizational Affiliation:
Division of Biotechnology, Department of Biology, Friedrich-Alexander-University Erlangen-Nuremberg, Henkestr. 91, D-91052 Erlangen, Germany.