Molecular mechanism of two nanobodies that inhibit PAI-1 activity reveals a modulation at distinct stages of the PAI-1/plasminogen activator interaction.
Sillen, M., Weeks, S.D., Zhou, X., Komissarov, A.A., Florova, G., Idell, S., Strelkov, S.V., Declerck, P.J.(2020) J Thromb Haemost 18: 681-692
- PubMed: 31858714
- DOI: https://doi.org/10.1111/jth.14716
- Primary Citation of Related Structures:
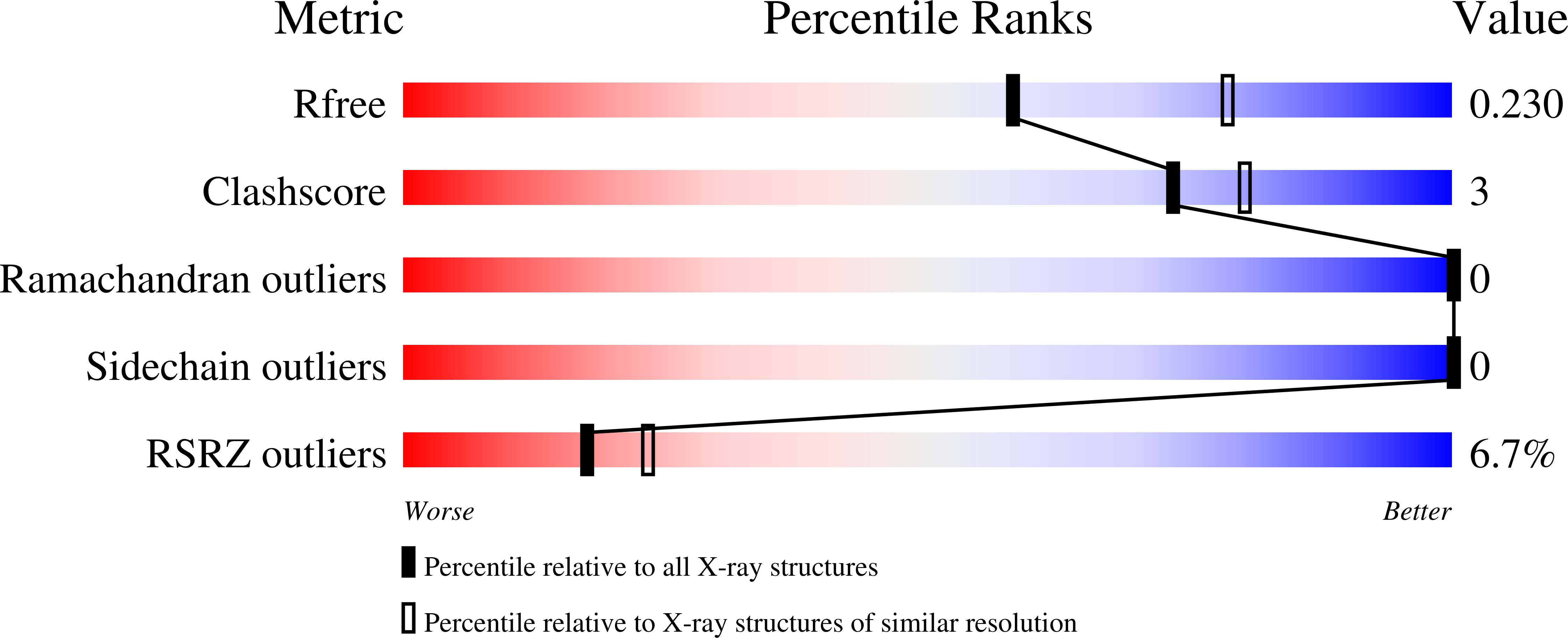
6GWN, 6GWP, 6GWQ - PubMed Abstract:
Plasminogen activator inhibitor-1 (PAI-1), a key inhibitor of plasminogen activators (PAs) tissue-type PA (tPA) and urokinase-type PA (uPA) plays a crucial role in many (patho)physiological processes (e.g., cardiovascular disease, tissue fibrosis) as well as in many age-related pathologies. Therefore, much effort has been put into the development of small molecule or antibody-based PAI-1 inhibitors. To elucidate the molecular mechanism of nanobody-induced PAI-1 inhibition. Here we present the first crystal structures of PAI-1 in complex with two neutralizing nanobodies (Nbs). These structures, together with biochemical and biophysical characterization, reveal that Nb VHH-2g-42 (Nb42) interferes with the initial PAI-1/PA complex formation, whereas VHH-2w-64 (Nb64) redirects the PAI-1/PA interaction to PAI-1 deactivation and regeneration of active PA. Furthermore, whereas vitronectin does not have an impact on the inhibitory effect of Nb42, it strongly potentiates the inhibitory effect of Nb64, which may contribute to a strong inhibitory potential of Nb64 in vivo. These findings illuminate the molecular mechanisms of PAI-1 inhibition. Nb42 and Nb64 can be used as starting points to engineer further improved antibody-based PAI-1 inhibitors or guide the rational design of small molecule inhibitors to treat a wide range of PAI-1-related pathophysiological conditions.
Organizational Affiliation:
Laboratory for Therapeutic and Diagnostic Antibodies, Department of Pharmaceutical and Pharmacological Sciences, KU Leuven, Leuven, Belgium.