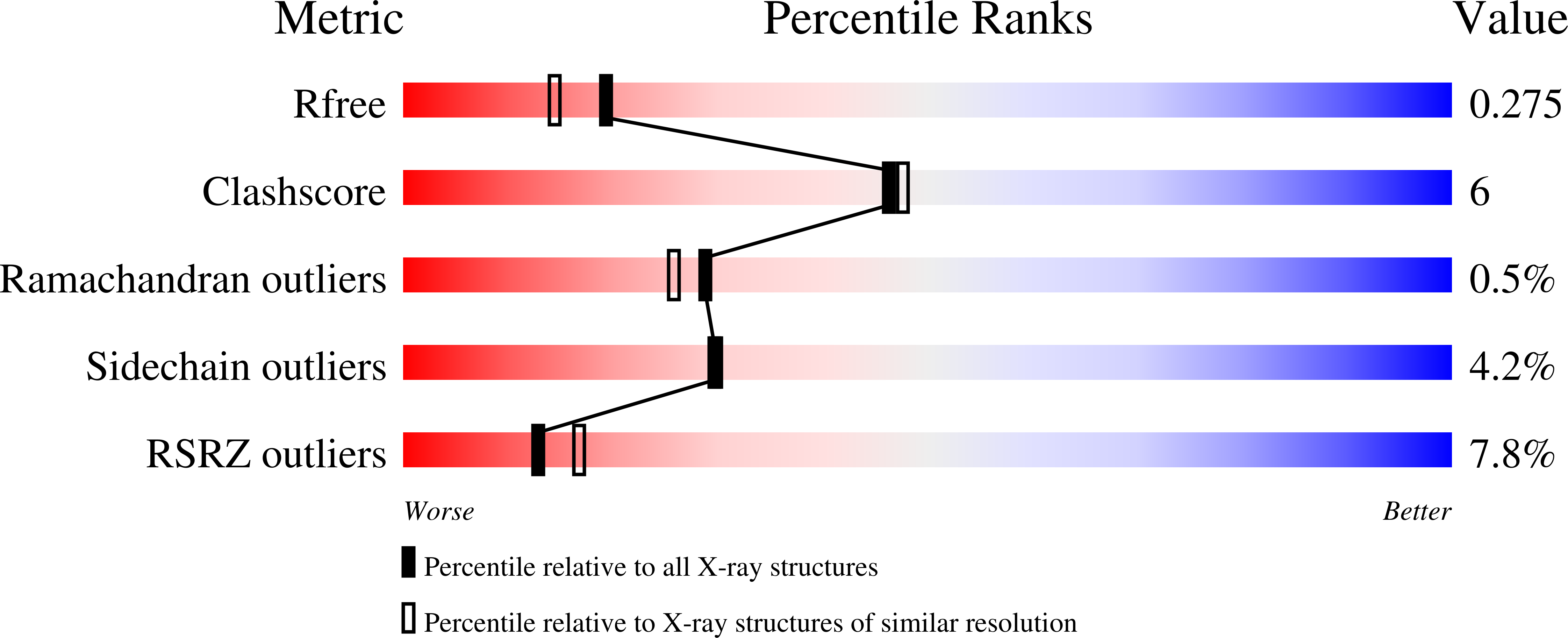
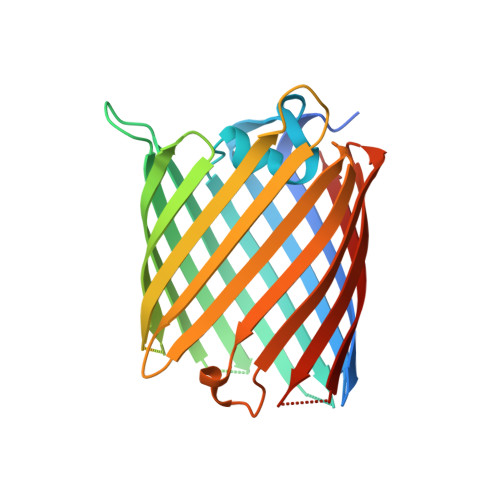
Crystal structure of the Acinetobacter baumannii outer membrane protein Omp33.
Abellon-Ruiz, J., Zahn, M., Basle, A., van den Berg, B.(2018) Acta Crystallogr D Struct Biol 74: 852-860
- PubMed: 30198896
- DOI: https://doi.org/10.1107/S205979831800904X
- Primary Citation of Related Structures:
6GIE - PubMed Abstract:
Acinetobacter baumannii is becoming a major threat to human health due to its multidrug resistance. This is owing in a large part to the low permeability of its outer membrane (OM), which prevents high internal antibiotic concentrations and makes antibiotic-resistance mechanisms more effective. To exploit OM channels as potential delivery vehicles for future antibiotics, structural information is required. One abundant OM protein in A. baumannii is Omp33. This protein has been reported to be important for the in vivo fitness and virulence of A. baumannii, but its structure is not known. Here, the X-ray crystal structure of Omp33 is reported at a resolution of 2.1 Å. Omp33 has a 14-β-stranded barrel without stable extracellular loop constrictions. Instead, an extended and unusual periplasmic turn connecting β-strands 2 and 3 is present, which folds into the pore lumen and completely blocks the aqueous channel. The Omp33 structure helps in understanding how A. baumannii OM proteins contribute to the low permeability of the cell envelope of this bacterium and suggests that Omp33 might function as a gated channel.
Organizational Affiliation:
Institute for Cell and Molecular Biosciences, The Medical School, Newcastle University, Newcastle upon Tyne NE2 4HH, England.