Trametes versicolor glutathione transferase Xi 3, a dual Cys-GST with catalytic specificities of both Xi and Omega classes.
Schwartz, M., Perrot, T., Deroy, A., Roret, T., Morel-Rouhier, M., Mulliert, G., Gelhaye, E., Favier, F., Didierjean, C.(2018) FEBS Lett 592: 3163-3172
- PubMed: 30112765
- DOI: https://doi.org/10.1002/1873-3468.13224
- Primary Citation of Related Structures:
6GC9, 6GCA, 6GCB, 6GCC, 6HTA - PubMed Abstract:
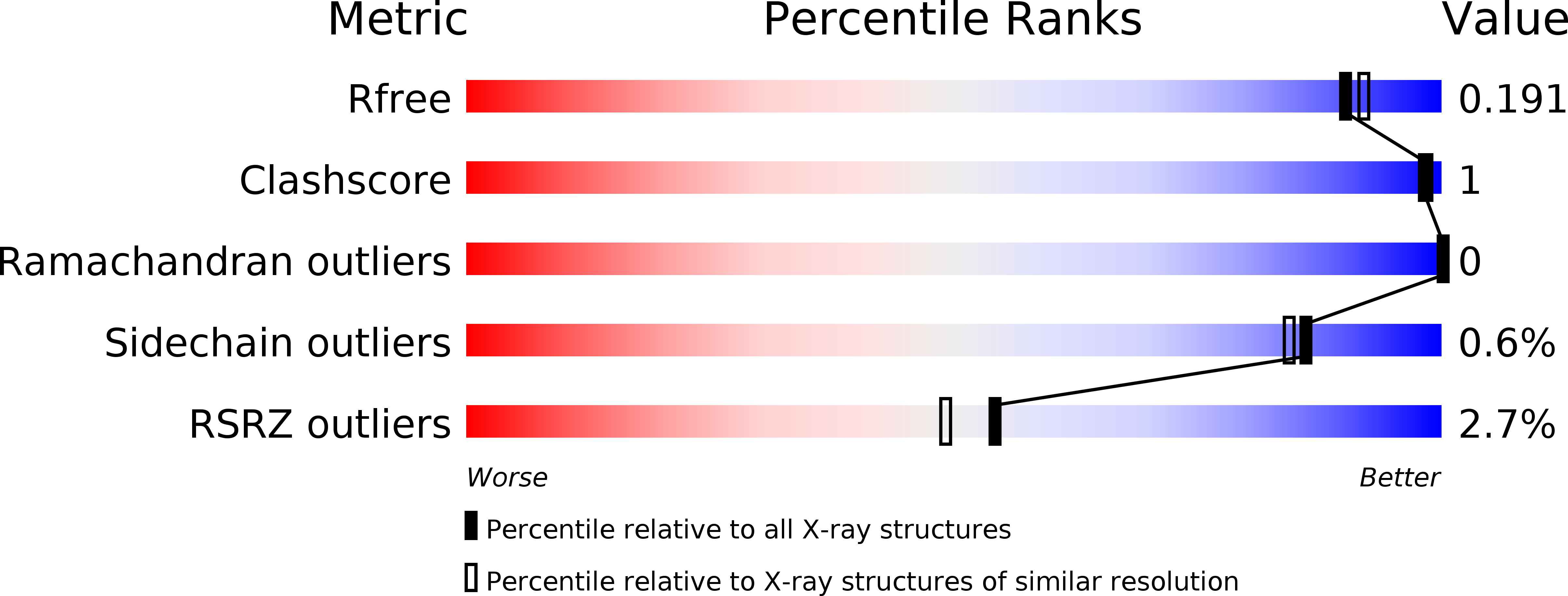
Glutathione transferases (GSTs) from the Xi and Omega classes have a catalytic cysteine residue, which gives them reductase activities. Until now, they have been assigned distinct substrates. While Xi GSTs specifically reduce glutathionyl-(hydro)quinones, Omega GSTs are specialized in the reduction of glutathionyl-acetophenones. Here, we present the biochemical and structural analysis of TvGSTX1 and TvGSTX3 isoforms from the wood-degrading fungus Trametes versicolor. TvGSTX1 reduces GS-menadione as expected, while TvGSTX3 reduces both Xi and Omega substrates. An in-depth structural analysis indicates a broader active site for TvGSTX3 due to specific differences in the nature of the residues situated in the C-terminal helix α9. This feature could explain the catalytic duality of TvGSTX3. Based on phylogenetic analysis, we propose that this duality might exist in saprophytic fungi and ascomycetes.
Organizational Affiliation:
CNRS, CRM2, Université de Lorraine, Nancy, France.