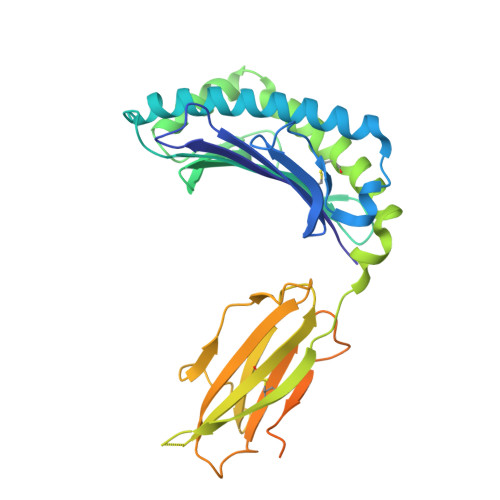
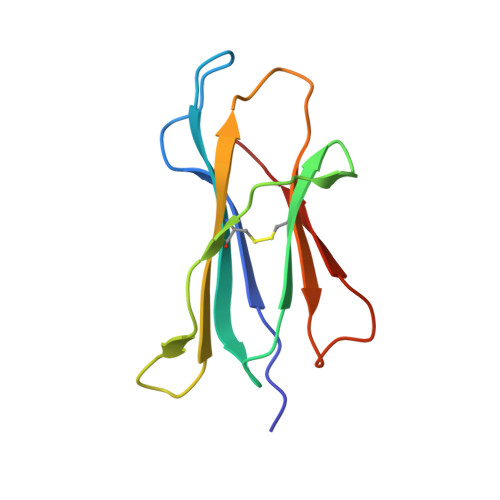
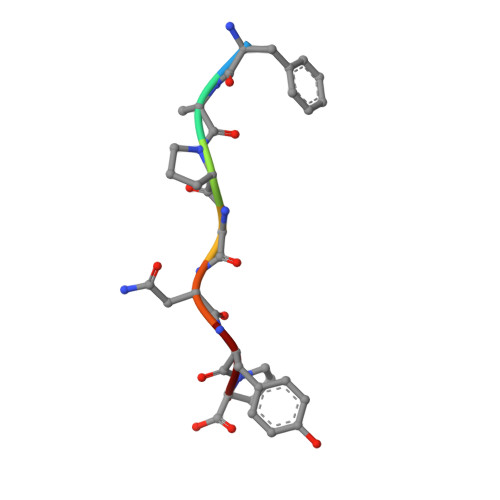
Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Hafstrand, I., Sayitoglu, E.C., Apavaloaei, A., Josey, B.J., Sun, R., Han, X., Pellegrino, S., Ozkazanc, D., Potens, R., Janssen, L., Nilvebrant, J., Nygren, P.A., Sandalova, T., Springer, S., Georgoudaki, A.M., Duru, A.D., Achour, A.(2019) Proc Natl Acad Sci U S A 116: 5055-5060
- PubMed: 30808808
- DOI: https://doi.org/10.1073/pnas.1807656116
- Primary Citation of Related Structures:
6GB5, 6GB6, 6GB7 - PubMed Abstract:
MHC-I epitope presentation to CD8 + T cells is directly dependent on peptide loading and selection during antigen processing. However, the exact molecular bases underlying peptide selection and binding by MHC-I remain largely unknown. Within the peptide-loading complex, the peptide editor tapasin is key to the selection of MHC-I-bound peptides. Here, we have determined an ensemble of crystal structures of MHC-I in complex with the peptide exchange-associated dipeptide GL, as well as the tapasin-associated scoop loop, alone or in combination with candidate epitopes. These results combined with mutation analyses allow us to propose a molecular model underlying MHC-I peptide selection by tapasin. The N termini of bound peptides most probably bind first in the N-terminal and middle region of the MHC-I peptide binding cleft, upon which the peptide C termini are tested for their capacity to dislodge the tapasin scoop loop from the F pocket of the MHC-I cleft. Our results also indicate important differences in peptide selection between different MHC-I alleles.
Organizational Affiliation:
Science for Life Laboratory, Department of Medicine Solna, Karolinska Institute, and Division of Infectious Diseases, Karolinska University Hospital, Solna, SE-17176 Stockholm, Sweden.