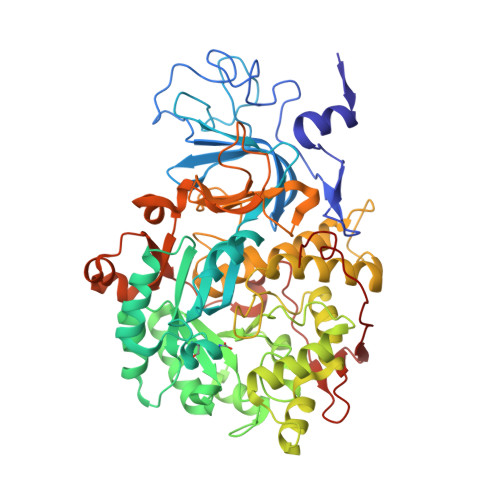
The structure of urease inactivated by Ag(i): a new paradigm for enzyme inhibition by heavy metals.
Mazzei, L., Cianci, M., Gonzalez Vara, A., Ciurli, S.(2018) Dalton Trans 47: 8240-8247
- PubMed: 29845996
- DOI: https://doi.org/10.1039/c8dt01190g
- Primary Citation of Related Structures:
6G48 - PubMed Abstract:
The nickel-dependent enzyme urease is a virulence factor for a large number of human pathogens, as well as a negative element for the efficiency of soil nitrogen fertilization for crop production. The use of urease inhibitors to contrast these effects requires the knowledge, at the molecular level, of their mode of action. Among these, silver is an efficient antimicrobial agent and an established inhibitor of this enzyme. The 1.91 Å resolution structure of Sporosarcina pasteurii urease inhibited by silver reveals the presence of two Ag(i) ions bound to the largely conserved triad αCys322/αHis323/αMet367: the first two residues are located on the mobile flap that is essential in modulating the size of the active site cavity and the position of key residues for enzyme catalysis, while αMet367 is on a loop facing the flap at the entrance of the active site cavity. The two Ag(i) ions are bridged by the thiolate Sγ atom of αCys322, and are coordinated, respectively, to the Nδ1 atom of the αHis323 imidazole ring and to the Sδ of αMet367. The binding of the Ag(i) ions at the edge of the active site channel supposedly blocks the movement of the flap, inhibiting the catalytic activity of urease. The structure of the silver-inhibited urease allows us to understand and rationalise all previously acquired kinetic and calorimetric data on this phenomenon, but also provides the details of how silver can exert its antimicrobial action with respect to ureolytic bacteria, a step forward against antibiotic-resistant pathogens.
Organizational Affiliation:
Laboratory of Bioinorganic Chemistry, Department of Pharmacy and Biotechnology, University of Bologna, Viale G. Fanin 40, I-40127 Bologna, Italy. stefano.ciurli@unibo.it.