N1-Substituted Quinoxaline-2,3-diones as Kainate Receptor Antagonists: X-ray Crystallography, Structure-Affinity Relationships, and in Vitro Pharmacology.
Pallesen, J., Mollerud, S., Frydenvang, K., Pickering, D.S., Bornholdt, J., Nielsen, B., Pasini, D., Han, L., Marconi, L., Kastrup, J.S., Johansen, T.N.(2019) ACS Chem Neurosci 10: 1841-1853
- PubMed: 30620174
- DOI: https://doi.org/10.1021/acschemneuro.8b00726
- Primary Citation of Related Structures:
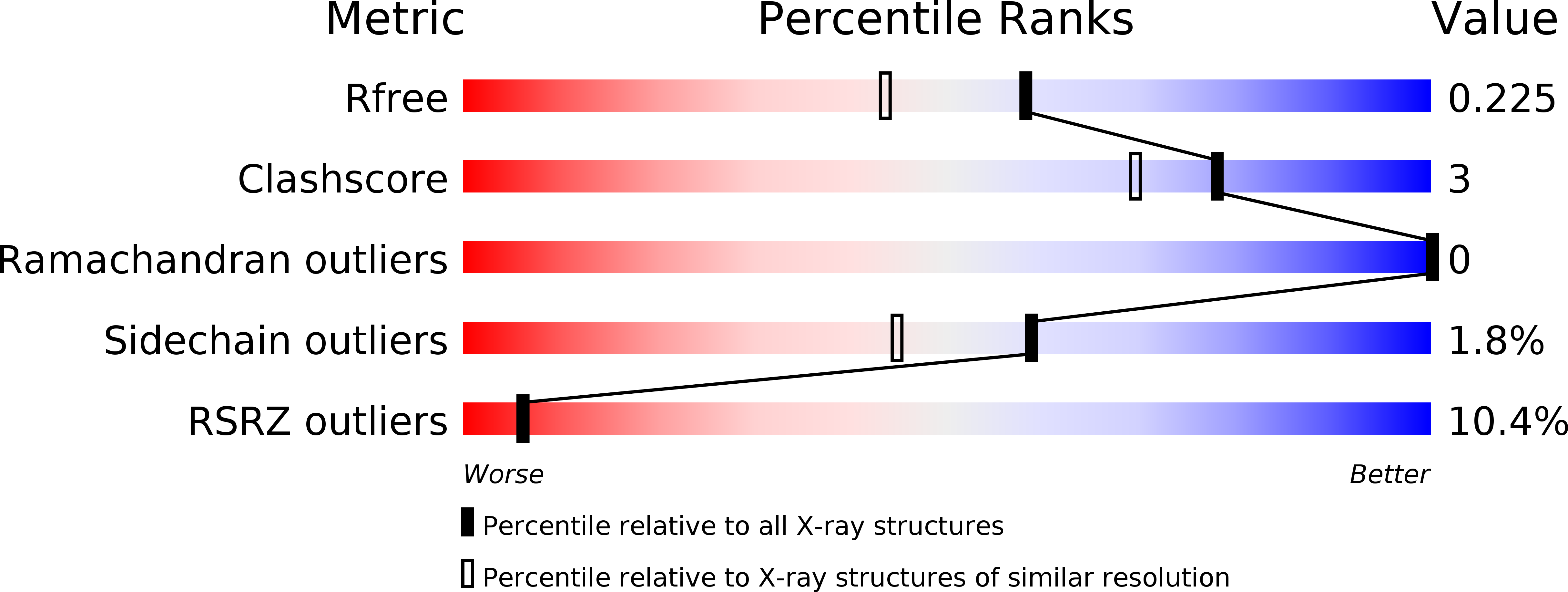
6FZ4 - PubMed Abstract:
Among the ionotropic glutamate receptors, the physiological role of kainate receptors is less well understood. Although ligands with selectivity toward the kainate receptor subtype GluK1 are available, tool compounds with selectivity at the remaining kainate receptor subtypes are sparse. Here, we have synthesized a series of quinoxaline-2,3-diones with substitutions in the N1-, 6-, and 7-position to investigate the structure-activity relationship (SAR) at GluK1-3 and GluK5. Pharmacological characterization at native and recombinant kainate and AMPA receptors revealed that compound 37 had a GluK3-binding affinity ( K i ) of 0.142 μM and 8-fold preference for GluK3 over GluK1. Despite lower binding affinity of 22 at GluK3 ( K i = 2.91 μM), its preference for GluK3 over GluK1 and GluK2 was >30-fold. Compound 37 was crystallized with the GluK1 ligand-binding domain to understand the SAR. The X-ray structure showed that 37 stabilized the protein in an open conformation, consistent with an antagonist binding mode.
Organizational Affiliation:
Department of Drug Design and Pharmacology, Faculty of Health and Medical Sciences , University of Copenhagen , DK-2100 Copenhagen , Denmark.