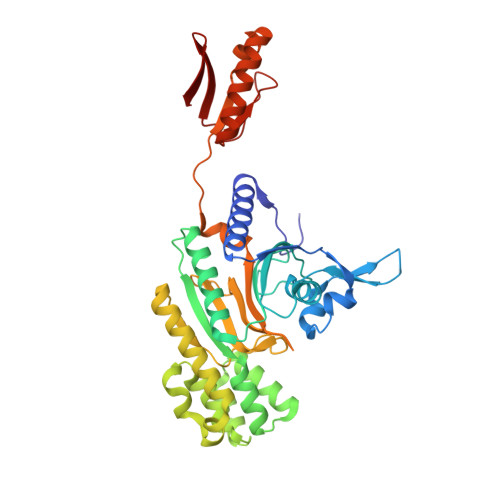
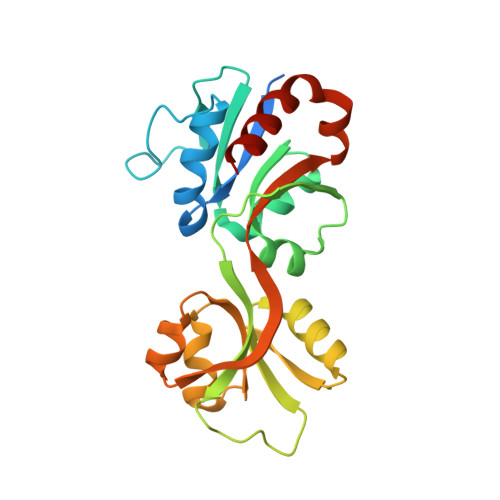
Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Alphey, M.S., Fisher, G., Hirschi, J.S., Stroek, R., Ge, Y., Gould, E.R., Czekster, C.M., Liu, H., Florence, G.J., Vetticatt, M.J., Naismith, J.H., da Silva, R.G.(2018) ACS Catal