Monoamine Biosynthesis via a Noncanonical Calcium-Activatable Aromatic Amino Acid Decarboxylase in Psilocybin Mushroom.
Torrens-Spence, M.P., Liu, C.T., Pluskal, T., Chung, Y.K., Weng, J.K.(2018) ACS Chem Biol 13: 3343-3353
- PubMed: 30484626
- DOI: https://doi.org/10.1021/acschembio.8b00821
- Primary Citation of Related Structures:
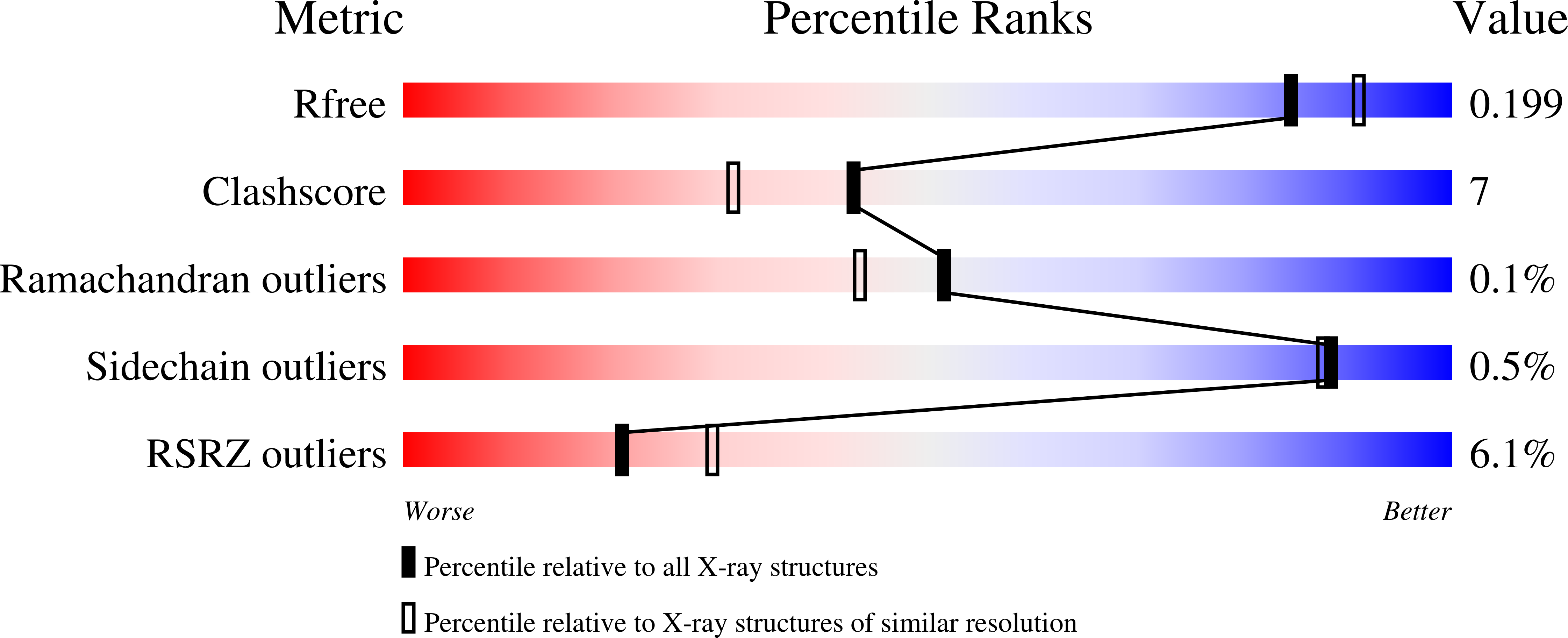
6EBN - PubMed Abstract:
Aromatic l-amino acid decarboxylases (AAADs) are a phylogenetically diverse group of enzymes responsible for the decarboxylation of aromatic amino acid substrates into their corresponding aromatic arylalkylamines. AAADs have been extensively studied in mammals and plants as they catalyze the first step in the production of neurotransmitters and bioactive phytochemicals, respectively. Unlike mammals and plants, the hallucinogenic psilocybin mushroom Psilocybe cubensis reportedly employs an unrelated phosphatidylserine-decarboxylase-like enzyme to catalyze l-tryptophan decarboxylation, the first step in psilocybin biosynthesis. To explore the origin of this chemistry in psilocybin mushroom, we generated the first de novo transcriptomes of P. cubensis and investigated several putative l-tryptophan-decarboxylase-like enzymes. We report the biochemical characterization of a noncanonical AAAD from P. cubensis ( PcncAAAD) that exhibits substrate permissiveness toward l-phenylalanine, l-tyrosine, and l-tryptophan, as well as chloro-tryptophan derivatives. The crystal structure of PcncAAAD revealed the presence of a unique C-terminal appendage domain featuring a novel double-β-barrel fold. This domain is required for PcncAAAD activity and regulates catalytic rate and thermal stability through calcium binding. PcncAAAD likely plays a role in psilocybin production in P. cubensis and offers a new tool for metabolic engineering of aromatic-amino-acid-derived natural products.
Organizational Affiliation:
Whitehead Institute for Biomedical Research , 455 Main Street , Cambridge , Massachusetts 02142 , United States.