Crystal structure of tryptophan synthase from M. tuberculosis - closed form with BRD6309 bound
Chang, C., Michalska, K., Maltseva, N.I., Jedrzejczak, R., McCarren, P., Nag, P.P., Joachimiak, A., Satchell, K.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tryptophan synthase alpha chain | 276 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: trpA, Rv1613, MTCY01B2.05 EC: 4.2.1.20 | 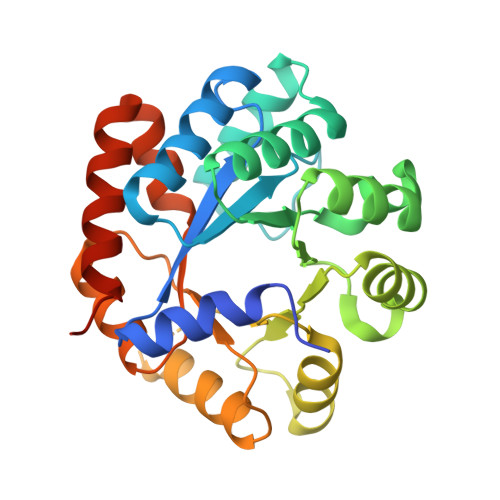 | |
UniProt | |||||
Find proteins for P9WFY1 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WFY1 Go to UniProtKB: P9WFY1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WFY1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tryptophan synthase beta chain | 410 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: trpB, Rv1612, MTCY01B2.04 EC: 4.2.1.20 | 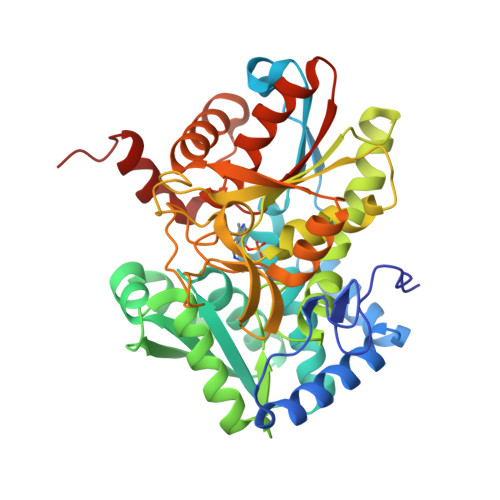 | |
UniProt | |||||
Find proteins for P9WFX9 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WFX9 Go to UniProtKB: P9WFX9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WFX9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HDJ Query on HDJ | AA [auth D], CB [auth H], L [auth B], RA [auth F] | (2R,3S,4R)-3-(2',6'-difluoro-4'-methyl[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile C18 H15 F3 N2 OZFKPHFHTNQIQS-XYJFISCASA-N |  | ||
| MLT Query on MLT | M [auth B] | D-MALATE C4 H6 O5 BJEPYKJPYRNKOW-UWTATZPHSA-N |  | ||
| MLA Query on MLA | I [auth A], W [auth C] | MALONIC ACID C3 H4 O4 OFOBLEOULBTSOW-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | AB [auth G] HA [auth D] HB [auth H] IA [auth D] IB [auth H] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | BB [auth G], J [auth A], O [auth B], Q [auth B], ZA [auth G] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| FMT Query on FMT | BA [auth D] CA [auth D] DA [auth D] DB [auth H] EA [auth D] | FORMIC ACID C H2 O2 BDAGIHXWWSANSR-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| LLP Query on LLP | B, D, F, H | L-PEPTIDE LINKING | C14 H22 N3 O7 P |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 134.66 | α = 90 |
| b = 157.944 | β = 90 |
| c = 166.078 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SCALEPACK | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-3000 | data reduction |
| HKL-3000 | phasing |