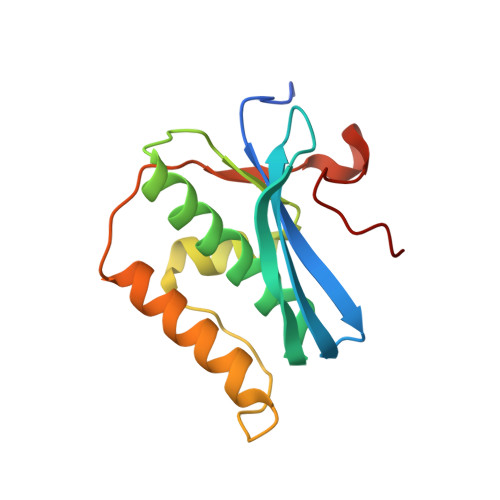
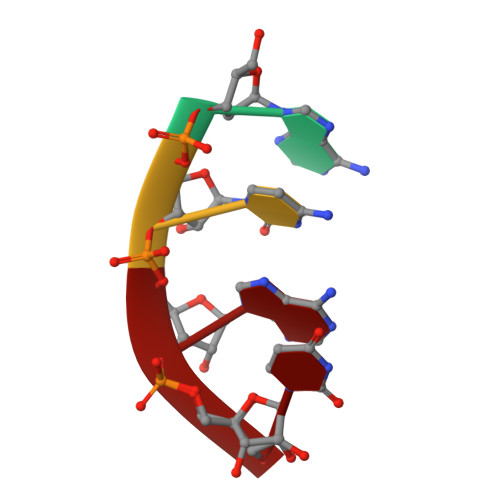
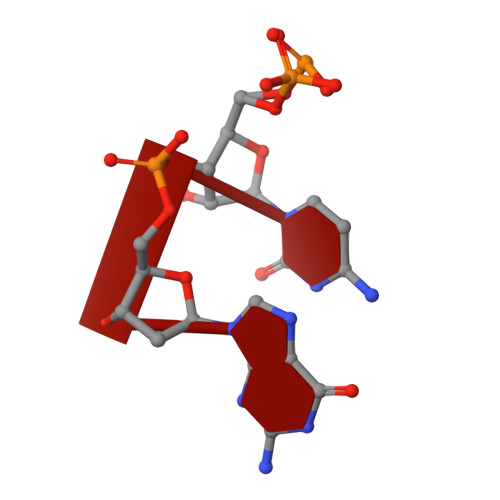
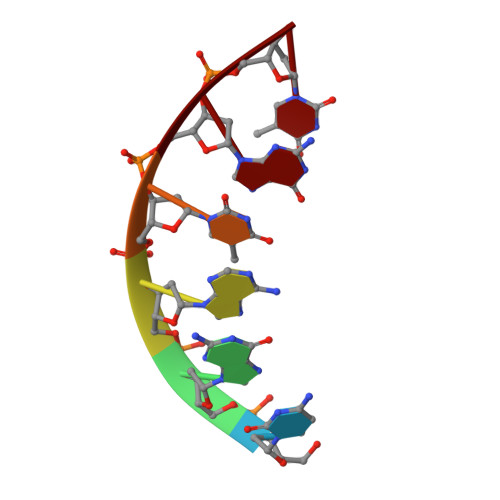
Cation trafficking propels RNA hydrolysis.
Samara, N.L., Yang, W.(2018) Nat Struct Mol Biol 25: 715-721
- PubMed: 30076410
- DOI: https://doi.org/10.1038/s41594-018-0099-4
- Primary Citation of Related Structures:
6DMN, 6DMV, 6DO8, 6DO9, 6DOA, 6DOB, 6DOC, 6DOD, 6DOE, 6DOF, 6DOG, 6DOH, 6DOI, 6DOJ, 6DOK, 6DOL, 6DOM, 6DON, 6DOO, 6DOP, 6DOQ, 6DOR, 6DOS, 6DOT, 6DOU, 6DOV, 6DOW, 6DOX, 6DOY, 6DOZ, 6DP0, 6DP1, 6DP2, 6DP3, 6DP4, 6DP5, 6DP6, 6DP7, 6DP8, 6DP9, 6DPA, 6DPB, 6DPC, 6DPD, 6DPE, 6DPF, 6DPG, 6DPH, 6DPI, 6DPJ - PubMed Abstract:
Catalysis by members of the RNase H superfamily of enzymes is generally believed to require only two Mg 2+ ions that are coordinated by active-site carboxylates. By examining the catalytic process of Bacillus halodurans RNase H1 in crystallo, however, we found that the two canonical Mg 2+ ions and an additional K + failed to align the nucleophilic water for RNA cleavage. Substrate alignment and product formation required a second K + and a third Mg 2+ , which replaced the first K + and departed immediately after cleavage. A third transient Mg 2+ has also been observed for DNA synthesis, but in that case it coordinates the leaving group instead of the nucleophile as in the case of the RNase H1 hydrolysis reaction. These transient cations have no contact with the enzymes. Other DNA and RNA enzymes that catalyze consecutive cleavage and strand-transfer reactions in a single active site may similarly require cation trafficking coordinated by the substrate.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), US National Institutes of Health (NIH), Bethesda, MD, USA.