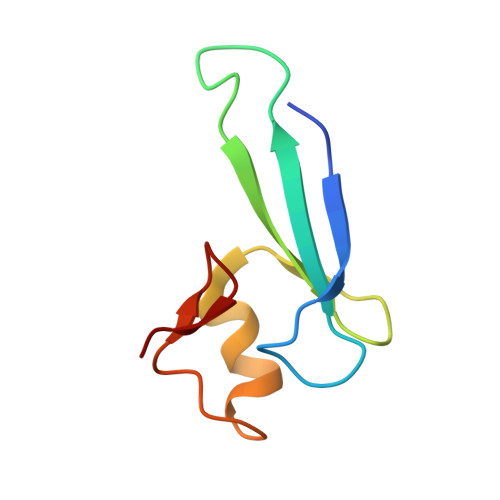
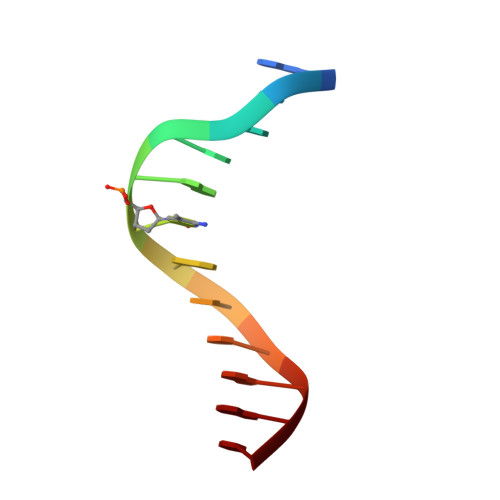
Structural analyses reveal that MBD3 is a methylated CG binder.
Liu, K., Lei, M., Wu, Z., Gan, B., Cheng, H., Li, Y., Min, J.(2019) FEBS J 286: 3240-3254
- PubMed: 30980593
- DOI: https://doi.org/10.1111/febs.14850
- Primary Citation of Related Structures:
6CC8, 6CCG, 6CEU, 6CEV - PubMed Abstract:
The MBD3, a methyl-CpG-binding domain (MBD)-containing protein, is a core subunit of the Mi-2/NuRD complex. Recent reports show that MBD3 recognizes both methylated CG (mCG)- and hydroxymethylated CG (hmCG)-containing DNA, with a preference for hmCG. However, whether the MBD3-MBD indeed has methyl-CG-binding ability is controversial. In this study, we provided the structural basis to support the ability of MBD3-MBD to bind mCG-containing DNA. We found that the MBD3-MBD bound to mCG-containing DNA through two conserved arginine fingers, and preferentially bound to mCG over hmCG, similar to other methyl-CpG-binding MBD proteins. Compared to its closest homolog MBD2, the tyrosine-to-phenylalanine substitution at Phe34 of MBD3 is responsible for a weaker mCG DNA binding ability. Based on the complex structure of MBD3-MBD with a nonpalindromic AmCGC DNA, we suggest that all the mCG-binding MBD domains can recognize mCG-containing DNA without orientation selectivity, consistent with our observations that the sequences outside the mCG dinucleotide do not affect mCG DNA binding significantly. DNA cytosine methylation is evolutionarily conserved in most metazoans, and most invertebrates have only one MBD gene, MBD2/3. We also looked into the mCG DNA binding ability of some invertebrates MBD2/3 and found that the conserved arginine fingers and a conserved structural fold are required for methylated DNA binding by MBD2/3-MBDs in invertebrates. Hence, our results demonstrate that mCG-binding arginine fingers embedded into a conserved structural fold are essential structural features for MBD2/3s binding to methylated DNA among metazoans.
Organizational Affiliation:
Hubei Key Laboratory of Genetic Regulation and Integrative Biology, School of Life Sciences, Central China Normal University, Wuhan, China.