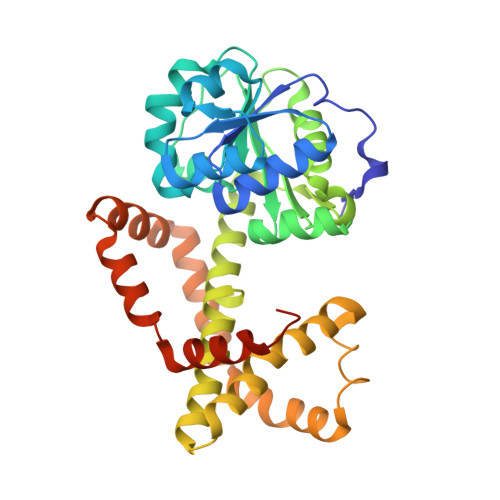
Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductosiomerase (KARI) inhibitor
Kandale, A., Patel, K.M., Zheng, S., You, L., Guddat, L.W., Schenk, G., Schembri, M.A., McGeary, R.P.To be published.