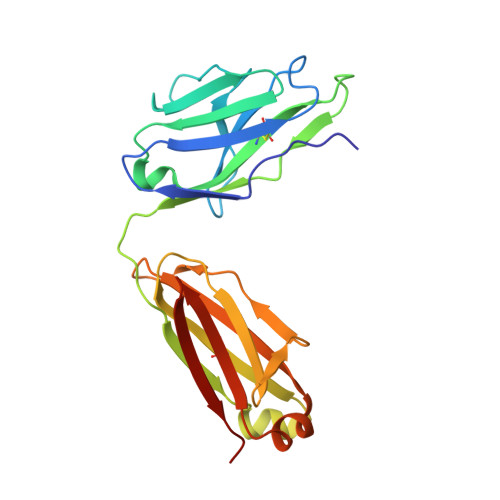
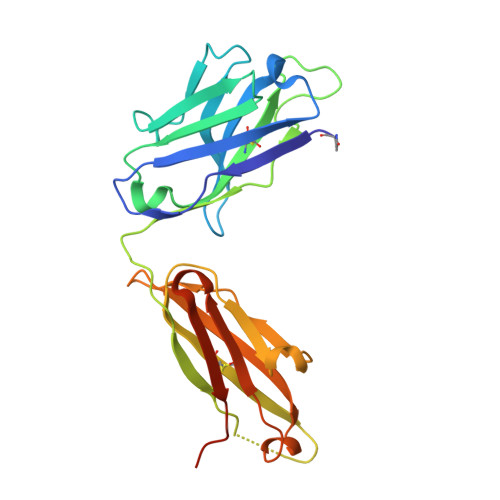
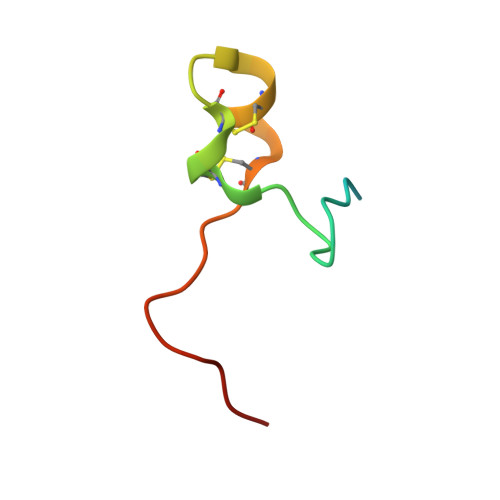
Structural basis for recognition of the central conserved region of RSV G by neutralizing human antibodies.
Jones, H.G., Ritschel, T., Pascual, G., Brakenhoff, J.P.J., Keogh, E., Furmanova-Hollenstein, P., Lanckacker, E., Wadia, J.S., Gilman, M.S.A., Williamson, R.A., Roymans, D., van 't Wout, A.B., Langedijk, J.P., McLellan, J.S.(2018) PLoS Pathog 14: e1006935-e1006935
- PubMed: 29509814
- DOI: https://doi.org/10.1371/journal.ppat.1006935
- Primary Citation of Related Structures:
6BLH, 6BLI - PubMed Abstract:
Respiratory syncytial virus (RSV) is a major cause of severe lower respiratory tract infections in infants and the elderly, and yet there remains no effective treatment or vaccine. The surface of the virion is decorated with the fusion glycoprotein (RSV F) and the attachment glycoprotein (RSV G), which binds to CX3CR1 on human airway epithelial cells to mediate viral attachment and subsequent infection. RSV G is a major target of the humoral immune response, and antibodies that target the central conserved region of G have been shown to neutralize both subtypes of RSV and to protect against severe RSV disease in animal models. However, the molecular underpinnings for antibody recognition of this region have remained unknown. Therefore, we isolated two human antibodies directed against the central conserved region of RSV G and demonstrated that they neutralize RSV infection of human bronchial epithelial cell cultures in the absence of complement. Moreover, the antibodies protected cotton rats from severe RSV disease. Both antibodies bound with high affinity to a secreted form of RSV G as well as to a peptide corresponding to the unglycosylated central conserved region. High-resolution crystal structures of each antibody in complex with the G peptide revealed two distinct conformational epitopes that require proper folding of the cystine noose located in the C-terminal part of the central conserved region. Comparison of these structures with the structure of fractalkine (CX3CL1) alone or in complex with a viral homolog of CX3CR1 (US28) suggests that RSV G would bind to CX3CR1 in a mode that is distinct from that of fractalkine. Collectively, these results build on recent studies demonstrating the importance of RSV G in antibody-mediated protection from severe RSV disease, and the structural information presented here should guide the development of new vaccines and antibody-based therapies for RSV.
Organizational Affiliation:
Department of Biochemistry and Cell Biology, Geisel School of Medicine at Dartmouth, Hanover, New Hampshire, United States of America.