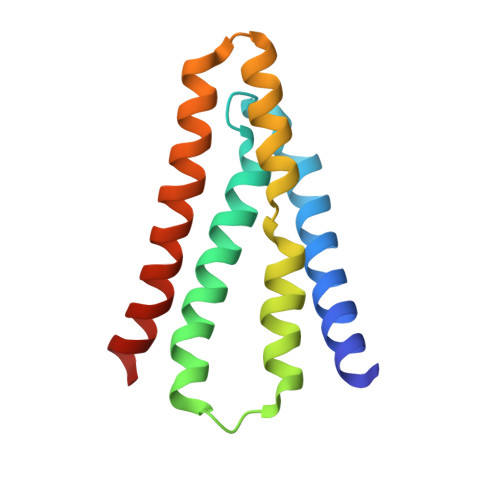
Molecular determinants of permeation in a fluoride-specific ion channel.
Last, N.B., Sun, S., Pham, M.C., Miller, C.(2017) Elife 6
- PubMed: 28952925
- DOI: https://doi.org/10.7554/eLife.31259
- Primary Citation of Related Structures:
6B24, 6B2A, 6B2B, 6B2D - PubMed Abstract:
Fluoride ion channels of the Fluc family combat toxicity arising from accumulation of environmental F - . Although crystal structures are known, the densely packed pore region has precluded delineation of the ion pathway. Here we chart out the Fluc pore and characterize its chemical requirements for transport. A ladder of H-bond donating residues creates a 'polar track' demarking the ion-conduction pathway. Surprisingly, while track polarity is well conserved, polarity is nonetheless functionally dispensable at several positions. A threonine at one end of the pore engages in vital interactions through its β-branched methyl group. Two critical central phenylalanines that directly coordinate F - through a quadrupolar-ion interaction cannot be functionally substituted by aromatic, non-polar, or polar sidechains. The only functional replacement is methionine, which coordinates F - through its partially positive γ-methylene in mimicry of phenylalanine's quadrupolar interaction. These results demonstrate the unusual chemical requirements for selectively transporting the strongly H-bonding F - anion.
Organizational Affiliation:
Department of Biochemistry, Howard Hughes Medical Institute, Brandeis University, Waltham, United States.