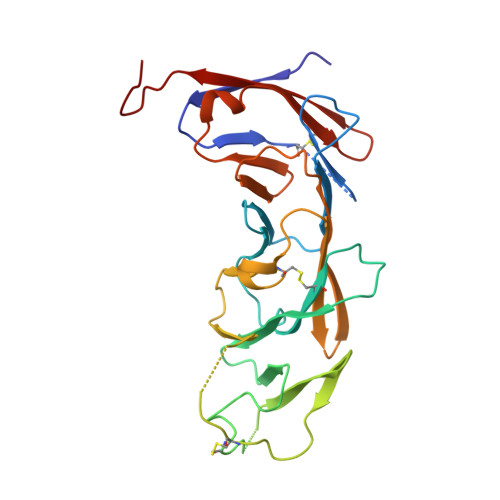
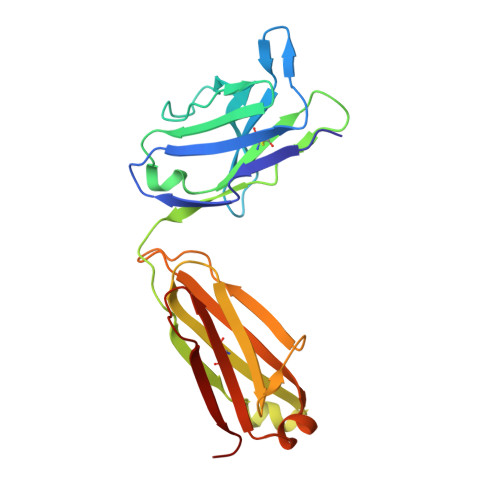
Crystal structure of B-cell co-receptor CD19 in complex with antibody B43 reveals an unexpected fold.
Teplyakov, A., Obmolova, G., Luo, J., Gilliland, G.L.(2018) Proteins 86: 495-500
- PubMed: 29490423
- DOI: https://doi.org/10.1002/prot.25485
- Primary Citation of Related Structures:
6AL4, 6AL5 - PubMed Abstract:
CD19 is a transmembrane protein expressed on malignant B cells, but not in other lineages or other tissues, which makes it an attractive target for monoclonal antibody-mediated immunotherapy. Anti-CD19 antibody B43 was utilized in a bispecific T-cell engager (BiTE) blinatumomab that demonstrated potency for the treatment of relapsed acute lymphoblastic leukemia. To gain insight into the mechanism of action of the antibody, the crystal structure of B43 Fab was determined in complex with CD19 and in the unbound form. The structure revealed the binding epitope, explained the lack of cross-reactivity toward non-human species, and suggested the key-and-lock mechanism of antigen recognition. Most unexpectedly, the structure revealed a unique molecular topology of CD19. Rather than a tandem of c-type immunoglobulin folds predicted from the amino acid sequence, the extracellular domain of CD19 exhibits an elongated β-sandwich formed by two immunoglobulin folds by swapping their C-terminal halves. This is the first structure of CD19, which has no sequence homologs.
Organizational Affiliation:
Janssen Research and Development, LLC, 1400 McKean Road, Spring House, Pennsylvania, 19477.