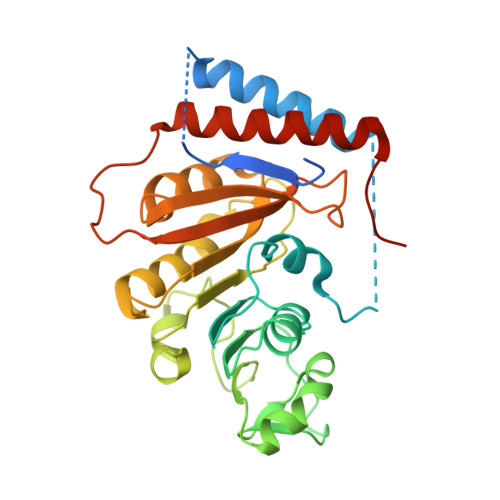
First insights into the structural features of Ebola virus methyltransferase activities.
Valle, C., Martin, B., Ferron, F., Roig-Zamboni, V., Desmyter, A., Debart, F., Vasseur, J.J., Canard, B., Coutard, B., Decroly, E.(2021) Nucleic Acids Res 49: 1737-1748
- PubMed: 33503246
- DOI: https://doi.org/10.1093/nar/gkaa1276
- Primary Citation of Related Structures:
6YU8 - PubMed Abstract:
The Ebola virus is a deadly human pathogen responsible for several outbreaks in Africa. Its genome encodes the 'large' L protein, an essential enzyme that has polymerase, capping and methyltransferase activities. The methyltransferase activity leads to RNA co-transcriptional modifications at the N7 position of the cap structure and at the 2'-O position of the first transcribed nucleotide. Unlike other Mononegavirales viruses, the Ebola virus methyltransferase also catalyses 2'-O-methylation of adenosines located within the RNA sequences. Herein, we report the crystal structure at 1.8 Å resolution of the Ebola virus methyltransferase domain bound to a fragment of a camelid single-chain antibody. We identified structural determinants and key amino acids specifically involved in the internal adenosine-2'-O-methylation from cap-related methylations. These results provide the first high resolution structure of an ebolavirus L protein domain, and the framework to investigate the effects of epitranscriptomic modifications and to design possible antiviral drugs against the Filoviridae family.
Organizational Affiliation:
AFMB, CNRS, Université Aix-Marseille, UMR 7257, Case 925, 163 Avenue de Luminy, 13288 Marseille Cedex 09, France.