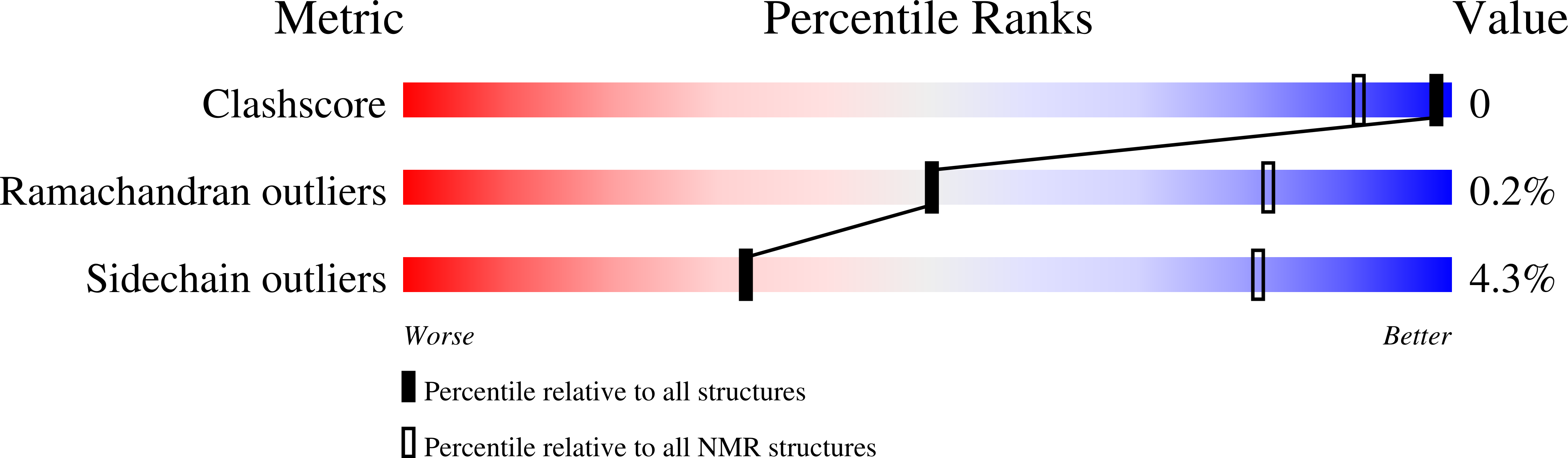Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Fuhrer, S., Kamenik, A.S., Zeindl, R., Nothegger, B., Hofer, F., Reider, N., Liedl, K.R., Tollinger, M.(2021) Sci Rep 11: 4173-4173
- PubMed: 33603065
- DOI: https://doi.org/10.1038/s41598-021-83705-z
- Primary Citation of Related Structures:
6Y3H, 6Y3I, 6Y3K, 6Y3L - PubMed Abstract:
A major proportion of allergic reactions to hazelnuts (Corylus avellana) are caused by immunologic cross-reactivity of IgE antibodies to pathogenesis-related class 10 (PR-10) proteins. Intriguingly, the four known isoforms of the hazelnut PR-10 allergen Cor a 1, denoted as Cor a 1.0401-Cor a 1.0404, share sequence identities exceeding 97% but possess different immunologic properties. In this work we describe the NMR solution structures of these proteins and provide an in-depth study of their biophysical properties. Despite sharing highly similar three-dimensional structures, the four isoforms exhibit remarkable differences regarding structural flexibility, hydrogen bonding and thermal stability. Our experimental data reveal an inverse relation between structural flexibility and IgE-binding in ELISA experiments, with the most flexible isoform having the lowest IgE-binding potential, while the isoform with the most rigid backbone scaffold displays the highest immunologic reactivity. These results point towards a significant entropic contribution to the process of antibody binding.
Organizational Affiliation:
Institute of Organic Chemistry and Center for Molecular Biosciences Innsbruck (CMBI), University of Innsbruck, Innrain 80/82, 6020, Innsbruck, Austria.