Discovery of CC-90011: A Potent and Selective Reversible Inhibitor of Lysine Specific Demethylase 1 (LSD1).
Kanouni, T., Severin, C., Cho, R.W., Yuen, N.Y., Xu, J., Shi, L., Lai, C., Del Rosario, J.R., Stansfield, R.K., Lawton, L.N., Hosfield, D., O'Connell, S., Kreilein, M.M., Tavares-Greco, P., Nie, Z., Kaldor, S.W., Veal, J.M., Stafford, J.A., Chen, Y.K.(2020) J Med Chem 63: 14522-14529
- PubMed: 33034194
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00978
- Primary Citation of Related Structures:
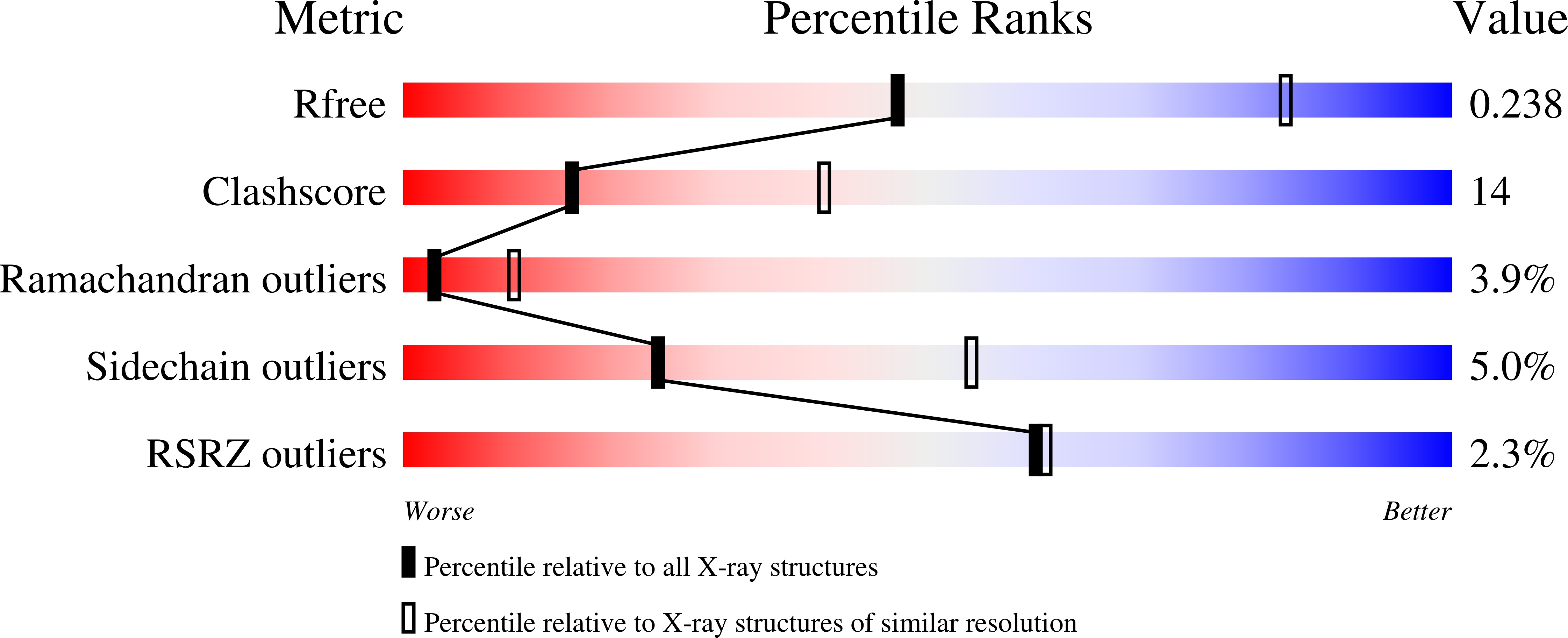
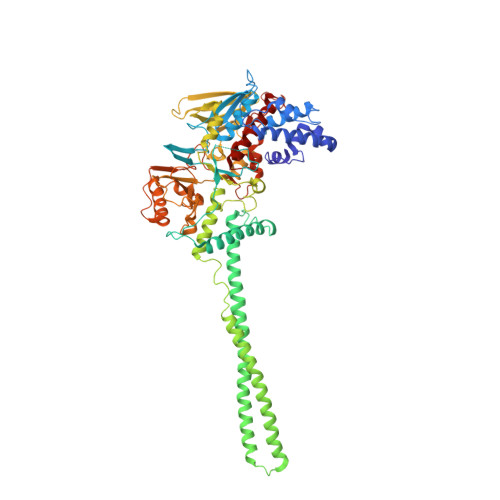
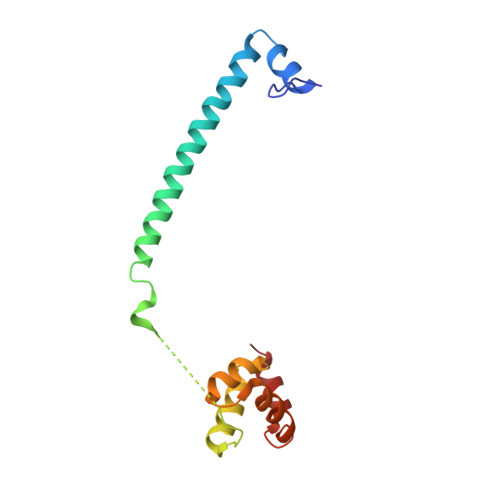
6W4K - PubMed Abstract:
Histone demethylase LSDl (KDMlA) belongs to the flavin adenine dinucleotide (FAD) dependent family of monoamine oxidases and is vital in regulation of mammalian biology. Dysregulation and overexpression of LSD1 are hallmarks of a number of human diseases, particularly cancers that are characterized as morphologically poorly differentiated. As such, inhibitors of LSD1 have potential to be beneficial as a cancer therapy. The most clinically advanced inhibitors of LSDl are covalent inhibitors derived from tranylcypromine (TCP). Herein, we report the discovery of a novel series of reversible and selective LSDl inhibitors. Exploration of structure-activity relationships (SARs) and optimization of ADME properties resulted in the identification of clinical candidate CC-90011. CC-90011 exhibits potent on-target induction of cellular differentiation in acute myeloid leukemia (AML) and small cell lung cancer (SCLC) cell lines, and antitumor efficacy in patient-derived xenograft (PDX) SCLC models. CC-90011 is currently in phase 2 trials in patients with first line, extensive stage SCLC (ClinicalTrials.gov identifier: NCT03850067).
Organizational Affiliation:
Fount Therapeutics, LLC, San Diego, California 92130, United States.