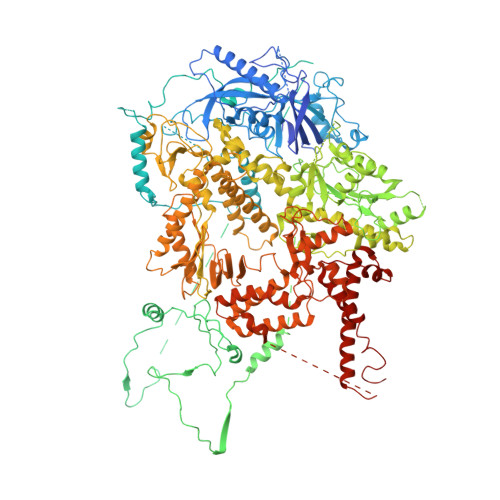
Structure and mechanism of B-family DNA polymerase zeta specialized for translesion DNA synthesis.
Malik, R., Kopylov, M., Gomez-Llorente, Y., Jain, R., Johnson, R.E., Prakash, L., Prakash, S., Ubarretxena-Belandia, I., Aggarwal, A.K.(2020) Nat Struct Mol Biol 27: 913-924
- PubMed: 32807989
- DOI: https://doi.org/10.1038/s41594-020-0476-7
- Primary Citation of Related Structures:
6V8P, 6V93 - PubMed Abstract:
DNA polymerase ζ (Polζ) belongs to the same B-family as high-fidelity replicative polymerases, yet is specialized for the extension reaction in translesion DNA synthesis (TLS). Despite its importance in TLS, the structure of Polζ is unknown. We present cryo-EM structures of the Saccharomyces cerevisiae Polζ holoenzyme in the act of DNA synthesis (3.1 Å) and without DNA (4.1 Å). Polζ displays a pentameric ring-like architecture, with catalytic Rev3, accessory Pol31' Pol32 and two Rev7 subunits forming an uninterrupted daisy chain of protein-protein interactions. We also uncover the features that impose high fidelity during the nucleotide-incorporation step and those that accommodate mismatches and lesions during the extension reaction. Collectively, we decrypt the molecular underpinnings of Polζ's role in TLS and provide a framework for new cancer therapeutics.
Organizational Affiliation:
Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, USA.