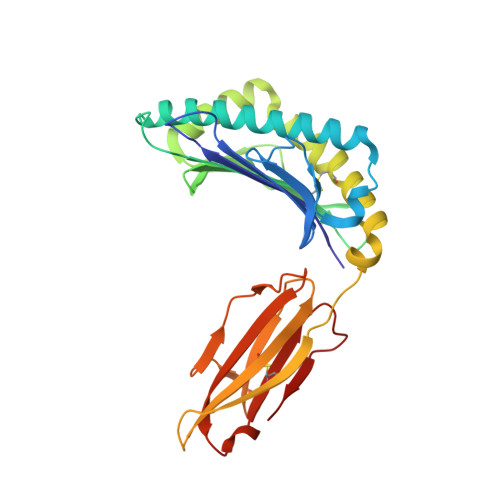
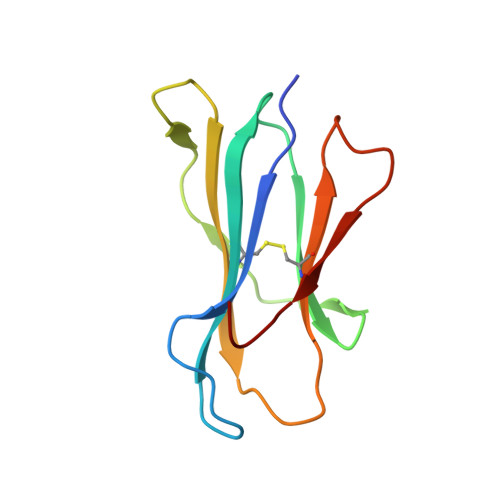
T cell receptor interactions with human leukocyte antigen govern indirect peptide selectivity for the cancer testis antigen MAGE-A4.
Coles, C.H., McMurran, C., Lloyd, A., Hock, M., Hibbert, L., Raman, M.C.C., Hayes, C., Lupardus, P., Cole, D.K., Harper, S.(2020) J Biol Chem 295: 11486-11494
- PubMed: 32532817
- DOI: https://doi.org/10.1074/jbc.RA120.014016
- Primary Citation of Related Structures:
6TRN, 6TRO - PubMed Abstract:
T cell-mediated immunity is governed primarily by T cell receptor (TCR) recognition of peptide-human leukocyte antigen (pHLA) complexes and is essential for immunosurveillance and disease control. This interaction is generally stabilized by interactions between the HLA surface and TCR germline-encoded complementarity-determining region (CDR) loops 1 and 2, whereas peptide selectivity is guided by direct interactions with the TCR CDR3 loops. Here, we solved the structure of a newly identified TCR in complex with a clinically relevant peptide derived from the cancer testis antigen melanoma antigen-A4 (MAGE-A4). The TCR bound pHLA in a position shifted toward the peptide's N terminus. This enabled the TCR to achieve peptide selectivity via an indirect mechanism, whereby the TCR sensed the first residue of the peptide through HLA residue Trp-167, which acted as a tunable gateway. Amino acid substitutions at peptide position 1 predicted to alter the HLA Trp-167 side-chain conformation abrogated TCR binding, indicating that this indirect binding mechanism is essential for peptide recognition. These findings extend our understanding of the molecular rules that underpin antigen recognition by TCRs and have important implications for the development of TCR-based therapies.
Organizational Affiliation:
Immunocore Ltd., Abingdon, United Kingdom.