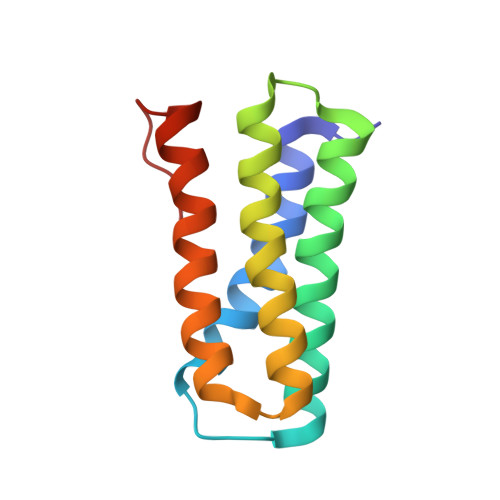
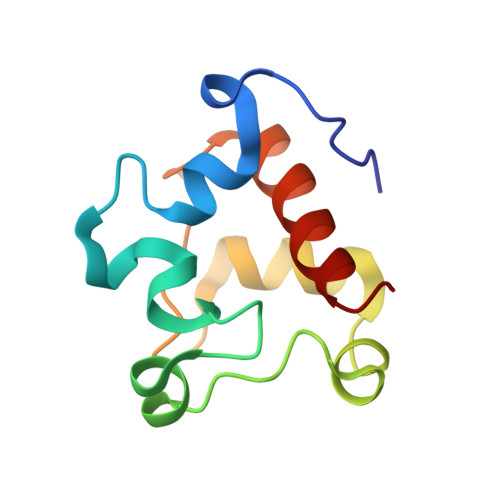
A nitric oxide-binding heterodimeric cytochromeccomplex from the anammox bacteriumKuenenia stuttgartiensisbinds to hydrazine synthase.
Akram, M., Reimann, J., Dietl, A., Menzel, A., Versantvoort, W., Kartal, B., Jetten, M.S.M., Barends, T.R.M.(2019) J Biol Chem 294: 16712-16728
- PubMed: 31548310
- DOI: https://doi.org/10.1074/jbc.RA119.008788
- Primary Citation of Related Structures:
6R6M, 6R6N, 6R6O - PubMed Abstract:
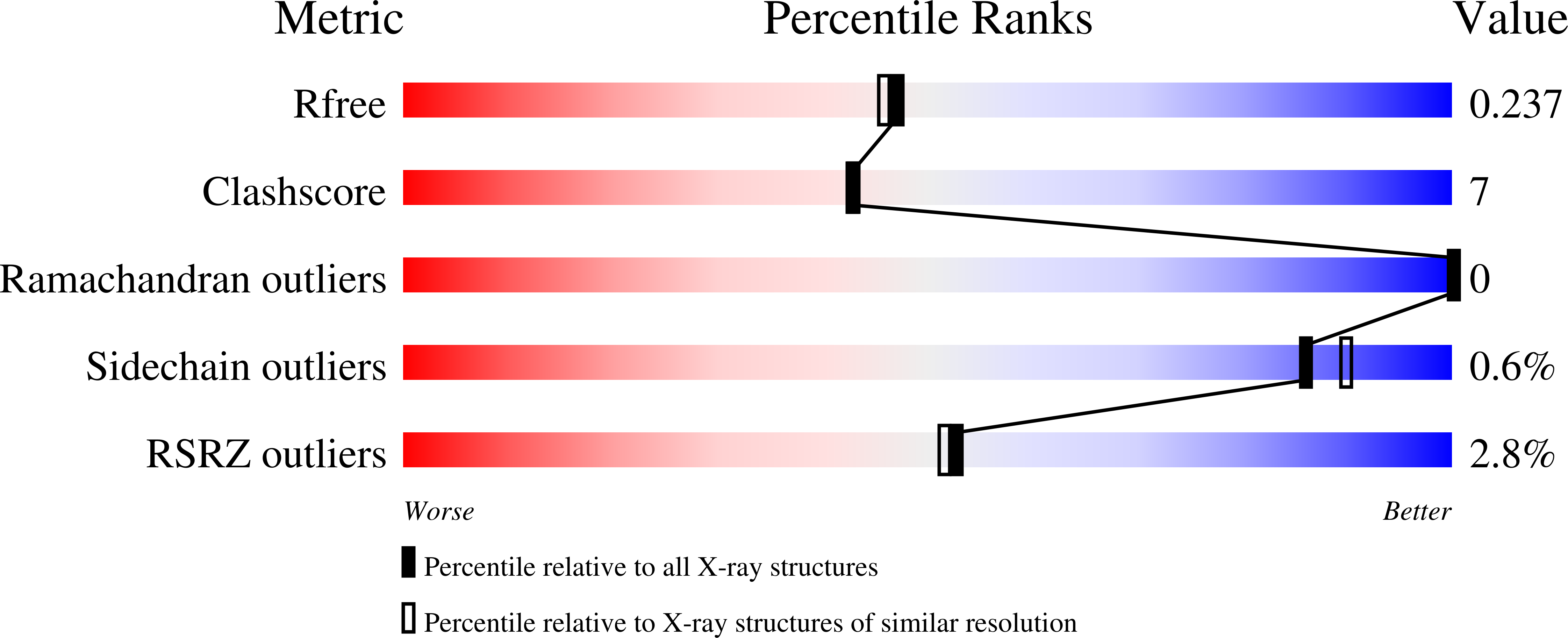
Anaerobic ammonium oxidation (anammox) is a microbial process responsible for significant nitrogen loss from the oceans and other ecosystems. The redox reactions at the heart of anammox are catalyzed by large multiheme enzyme complexes that rely on small cytochrome c proteins for electron shuttling. Among the most highly abundant of these cytochromes is a unique heterodimeric complex composed of class I and class II c -type cytochromes called NaxLS, which has distinctive biochemical and spectroscopic properties. Here, we present the 1.7 Å resolution crystal structure of this complex from the anammox organism Kuenenia stuttgartiensis (KsNaxLS). The structure reveals that the heme irons in each subunit exhibit a rare His/Cys ligation, which, as we show by substitution, causes the observed unusual spectral properties. Unlike its individual subunits, the KsNaxLS complex binds nitric oxide (NO) only at the distal heme side, forming 6cNO adducts. This is likely due to steric immobilization of the proximal heme-binding motifs upon complex formation, a finding that may be of functional relevance, because NO is an intermediate in the central anammox metabolism. Pulldown experiments with K. stuttgartiensis cell-free extract showed that the KsNaxLS complex binds specifically to one of the central anammox enzyme complexes, hydrazine synthase, which uses NO as one of its substrates. It is therefore possible that the KsNaxLS complex plays a role in binding the volatile NO to retain it in the cell for transfer to hydrazine synthase. Alternatively, we propose that KsNaxLS may shuttle electrons to this enzyme complex.
Organizational Affiliation:
Department of Biomolecular Mechanisms, Max Planck Institute for Medical Research, Heidelberg 69120, Germany.