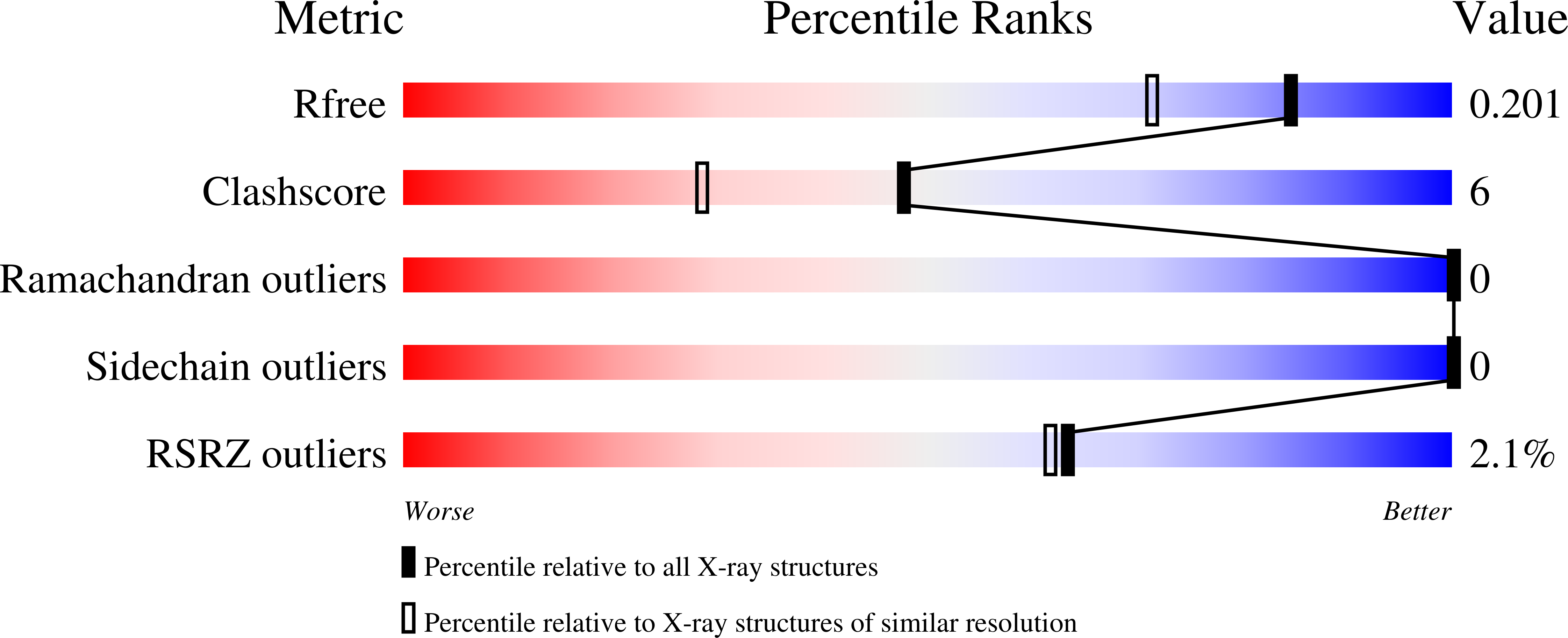
Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Birkinshaw, R.W., Gong, J.N., Luo, C.S., Lio, D., White, C.A., Anderson, M.A., Blombery, P., Lessene, G., Majewski, I.J., Thijssen, R., Roberts, A.W., Huang, D.C.S., Colman, P.M., Czabotar, P.E.(2019) Nat Commun 10: 2385-2385
- PubMed: 31160589
- DOI: https://doi.org/10.1038/s41467-019-10363-1
- Primary Citation of Related Structures:
6O0K, 6O0L, 6O0M, 6O0O, 6O0P - PubMed Abstract:
Venetoclax is a first-in-class cancer therapy that interacts with the cellular apoptotic machinery promoting apoptosis. Treatment of patients suffering chronic lymphocytic leukaemia with this BCL-2 antagonist has revealed emergence of a drug-selected BCL-2 mutation (G101V) in some patients failing therapy. To understand the molecular basis of this acquired resistance we describe the crystal structures of venetoclax bound to both BCL-2 and the G101V mutant. The pose of venetoclax in its binding site on BCL-2 reveals small but unexpected differences as compared to published structures of complexes with venetoclax analogues. The G101V mutant complex structure and mutant binding assays reveal that resistance is acquired by a knock-on effect of V101 on an adjacent residue, E152, with venetoclax binding restored by a E152A mutation. This provides a framework for considering analogues of venetoclax that might be effective in combating this mutation.
Organizational Affiliation:
Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, VIC, 3052, Australia. birkinshaw.r@wehi.edu.au.