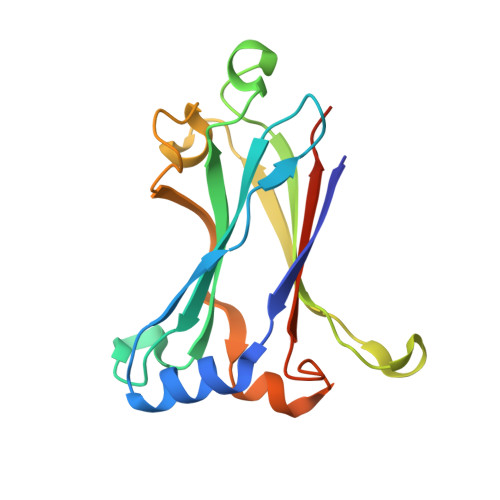
Binding and Enhanced Binding between Key Immunity Proteins TRAF6 and TIFA.
Huang, W.C., Liao, J.H., Hsiao, T.C., Wei, T.W., Maestre-Reyna, M., Bessho, Y., Tsai, M.D.(2019) Chembiochem 20: 140-146
- PubMed: 30378729
- DOI: https://doi.org/10.1002/cbic.201800436
- Primary Citation of Related Structures:
5ZUJ, 6A33 - PubMed Abstract:
Human tumor necrosis factor receptor associated factor (TRAF)-interacting protein, with a forkhead-associated domain (TIFA), is a key regulator of NF-κB activation. It also plays a key role in the activation of innate immunity in response to bacterial infection, through heptose 1,7-bisphosphate (HBP); a metabolite of lipopolysaccharide (LPS). However, the mechanism of TIFA function is largely unexplored, except for the suggestion of interaction with TRAF6. Herein, we provide evidence for direct binding, albeit weak, between TIFA and the TRAF domain of TRAF6, and it is shown that the binding is enhanced for a rationally designed double mutant, TIFA S174Q/M179D. Enhanced binding was also demonstrated for endogenous full-length TRAF6. Furthermore, the structures of the TRAF domain complexes with the consensus TRAF-binding peptides from the C terminus of wild-type and S174Q/M179D mutant TIFA, showing salt-bridge formation between residues 177-181 of TIFA and the binding pocket residues of the TRAF domain, were solved. Taken together, the results provide direct evidence and a structural basis for the TIFA-TRAF6 interaction, and show how this important biological function can be modulated.
Organizational Affiliation:
Institute of Biological Chemistry, Academia Sinica, 128 Academia Road Sec. 2, Nankang, Taipei, 115, Taiwan.