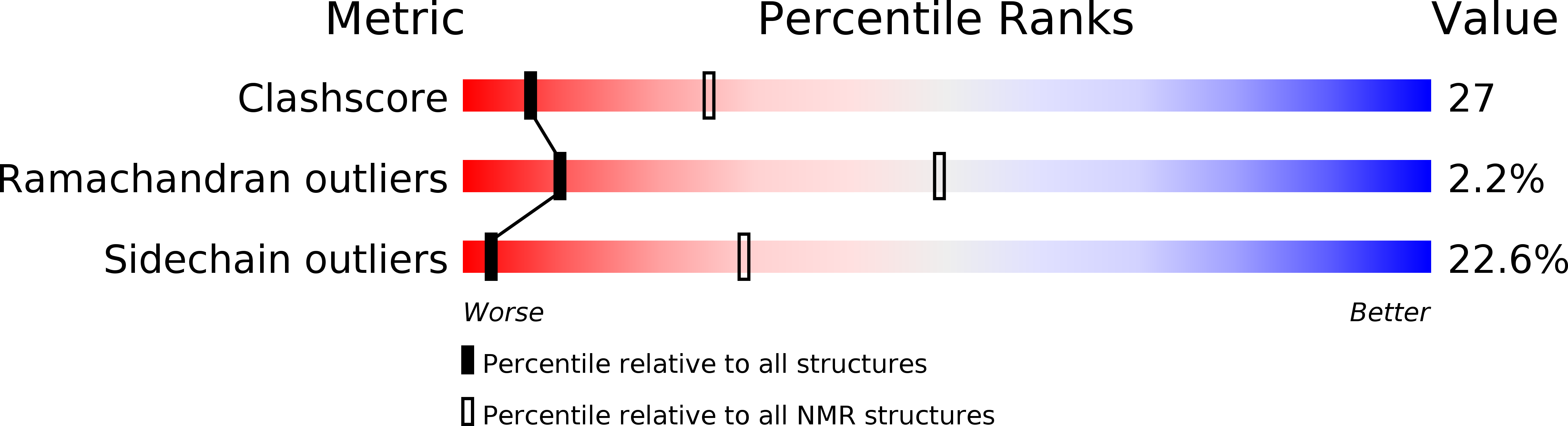Molecular Basis for Allosteric Inhibition of GTP-Bound H-Ras Protein by a Small-Molecule Compound Carrying a Naphthalene Ring
Matsumoto, S., Hiraga, T., Hayashi, Y., Yoshikawa, Y., Tsuda, C., Araki, M., Neya, M., Shima, F., Kataoka, T.(2018) Biochemistry 57: 5350-5358
- PubMed: 30141910
- DOI: https://doi.org/10.1021/acs.biochem.8b00680
- Primary Citation of Related Structures:
5ZC6 - PubMed Abstract:
The ras oncogene products (H-Ras, K-Ras, and N-Ras) have been regarded as some of the most promising targets for anticancer drug discovery because their activating mutations are frequently found in human cancers. Nonetheless, molecular targeted therapy for them is currently unavailable. Here, we report the discovery of a small-molecule compound carrying a naphthalene ring, named KBFM123, which binds to the GTP-bound form of H-Ras. The solution structure of its complex with the guanosine 5'-(β,γ-imide) triphosphate-bound form of H-RasT35S (H-RasT35S·GppNHp) indicates that the naphthalene ring of KBFM123 interacts directly with a hydrophobic pocket located between switch I and switch II and allosterically inhibits the effector interaction by inducing conformational changes in switch I and its flanking region in strand β2, which are directly involved in recognition of the effector molecules, including c-Raf-1. In particular, Asp38 of H-Ras, a crucial residue for the interaction with c-Raf-1 via the formation of a salt bridge with Arg89 of the Ras-binding domain (RBD) of c-Raf-1, shows a drastic conformational change: its side chain orients toward the opposite direction. Consistent with these results, KBFM123 exhibits an activity to inhibit, albeit weakly, the association of H-RasG12V·GppNHp with the c-Raf-1 RBD. The binding of the naphthalene ring to the hydrophobic pocket of H-RasT35S·GppNHp is further supported by nuclear magnetic resonance analyses showing that two other naphthalene-containing compounds with distinct structures also exhibit similar binding properties with KBFM123. These results indicate that the naphthalene ring could become a promising scaffold for the development of Ras inhibitors.
Organizational Affiliation:
Division of Molecular Biology, Department of Biochemistry and Molecular Biology , Kobe University Graduate School of Medicine , 7-5-1 Kusunoki-cho , Chuo-ku, Kobe 650-0017 , Japan.