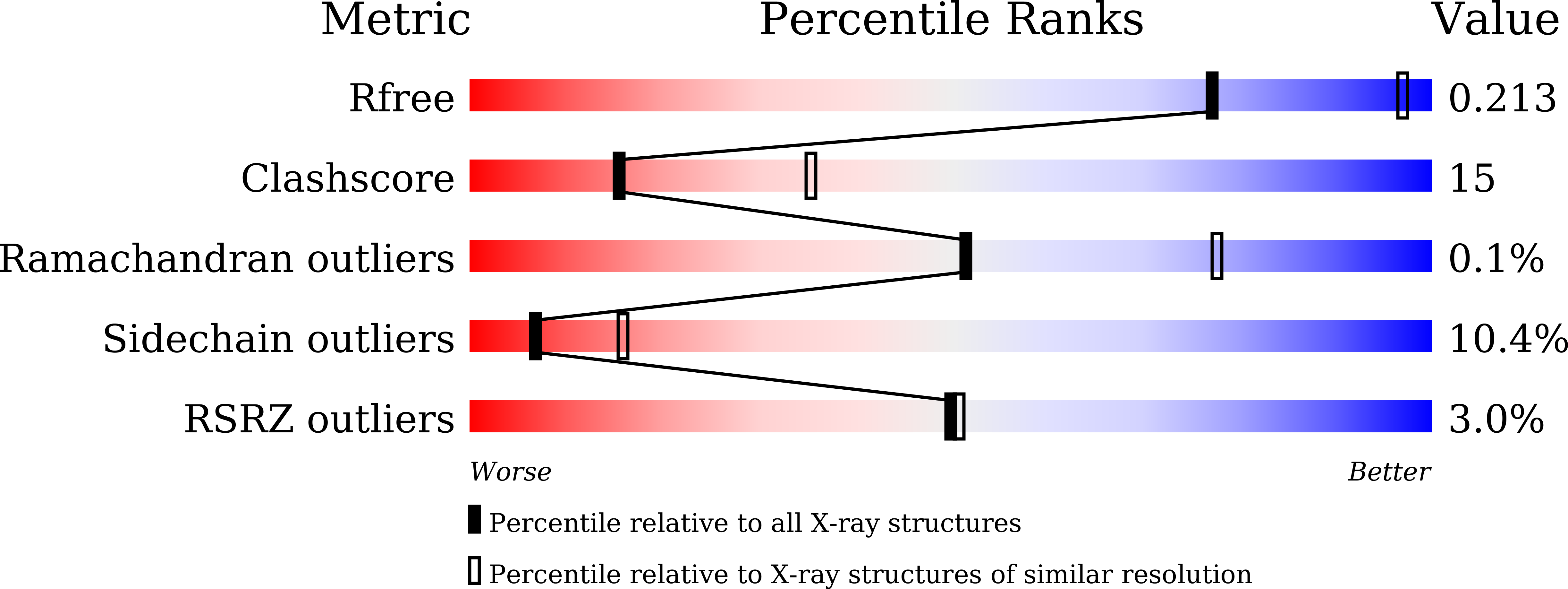
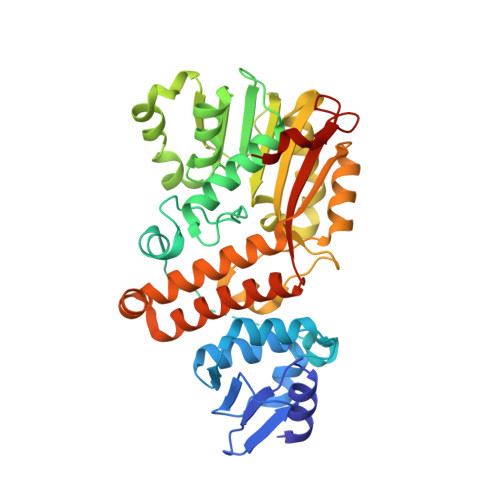
Crystal structure of bacterial cyclopropane-fatty-acyl-phospholipid synthase with phospholipid.
Ma, Y., Pan, C., Wang, Q.(2019) J Biochem 166: 139-147
- PubMed: 30828715
- DOI: https://doi.org/10.1093/jb/mvz018
- Primary Citation of Related Structures:
5Z9O - PubMed Abstract:
The lipids containing cyclopropane-fatty-acid (CFA) protect bacteria from adverse conditions such as acidity, freeze-drying desiccation and exposure to pollutants. CFA is synthesized when cyclopropane-fatty-acyl-phospholipid synthase (CFA synthase, CFAS) transfers a methylene group from S-adenosylmethionine (SAM) across the cis double bonds of unsaturated fatty acyl chains. Here, we reported a 2.7-Å crystal structure of CFAS from Lactobacillus acidophilus. The enzyme is composed of N- and C-terminal domain, which belong to the sterol carrier protein and methyltransferase superfamily, respectively. A phospholipid in the substrate binding site and a bicarbonate ion (BCI) acting as a general base in the active site were discovered. To elucidate the mechanism, a docking experiment using CFAS from L. acidophilus and SAM was carried out. The analysis of this structure demonstrated that three groups, the carbons from the substrate, the BCI and the methyl of S(CHn)3 group, were close enough to form a cyclopropane ring with the help of amino acids in the active site. Therefore, the structure supports the hypothesis that CFAS from L. acidophilus catalyzes methyl transfer via a carbocation mechanism. These findings provide a structural basis to more deeply understand enzymatic cyclopropanation.
Organizational Affiliation:
School of Bioengineering, Jingchu University of Technology, Jingmen, China.