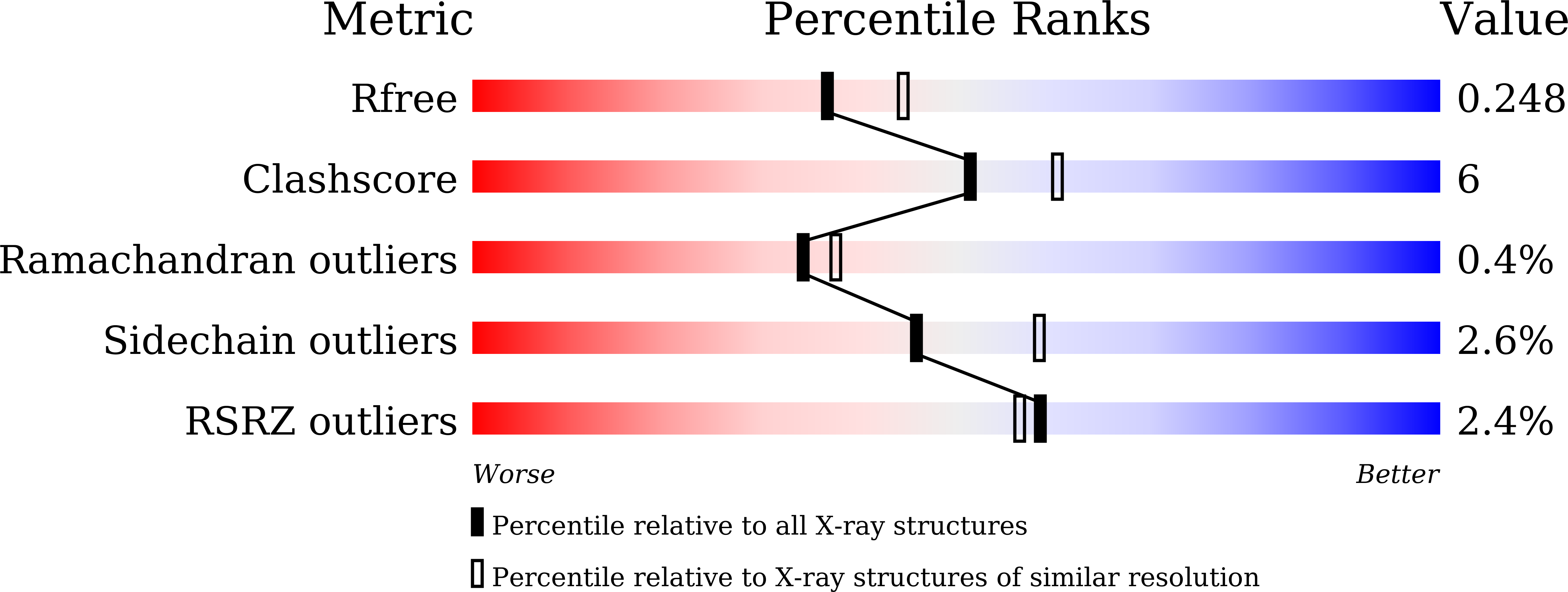
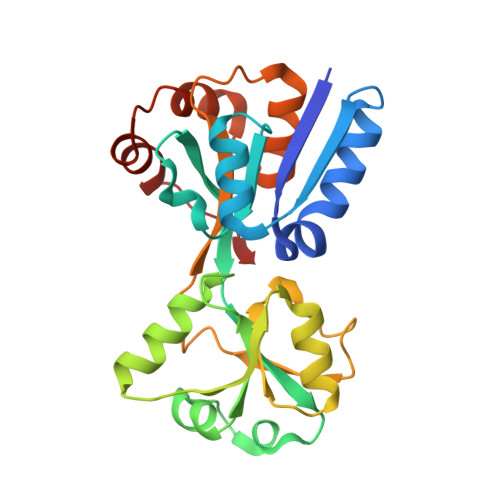
Molecular insights into the master regulator CysB-mediated bacterial virulence in Pseudomonas aeruginosa.
Song, Y., Yang, C., Chen, G., Zhang, Y., Seng, Z., Cai, Z., Zhang, C., Yang, L., Gan, J., Liang, H.(2019) Mol Microbiol 111: 1195-1210
- PubMed: 30618115
- DOI: https://doi.org/10.1111/mmi.14200
- Primary Citation of Related Structures:
5Z4Y, 5Z4Z, 5Z50 - PubMed Abstract:
Pseudomonas aeruginosa is a major pathogen that causes serious acute and chronic infections in humans. The type III secretion system (T3SS) is an important virulence factor that plays essential roles in acute infections. However, the regulatory mechanisms of T3SS are not fully understood. In this study, we found that the deletion of cysB reduced the T3SS gene expression and swarming motility but enhanced biofilm formation. In a mouse acute pneumonia model, mutation of cysB decreased the average bacterial load compared to that of the wild-type strain. Further experiments demonstrated that CysB contributed to the reduced T3SS gene expression and bacterial pathogenesis by directly regulating the sensor kinase RetS. We also performed crystallographic studies of PaCysB. The overall fold of PaCysB NTD domain is similar to other LysR superfamily proteins and structural superposition revealed one possible DNA-binding model for PaCysB. Structural comparison also revealed great flexibility of the PaCysB RD domain, which may play an important role in bending and transcriptional regulation of target DNA. Taken together, these results expand our current understanding of the complex regulatory networks of T3SS and RetS pathways. The crystal structure of CysB provides new insights for studying the function of its homologs in other bacterial species.
Organizational Affiliation:
Key Laboratory of Resources Biology and Biotechnology in Western China, Ministry of Education, College of Life Sciences, Northwest University, Xi'an, Shaanxi, 710069, China.