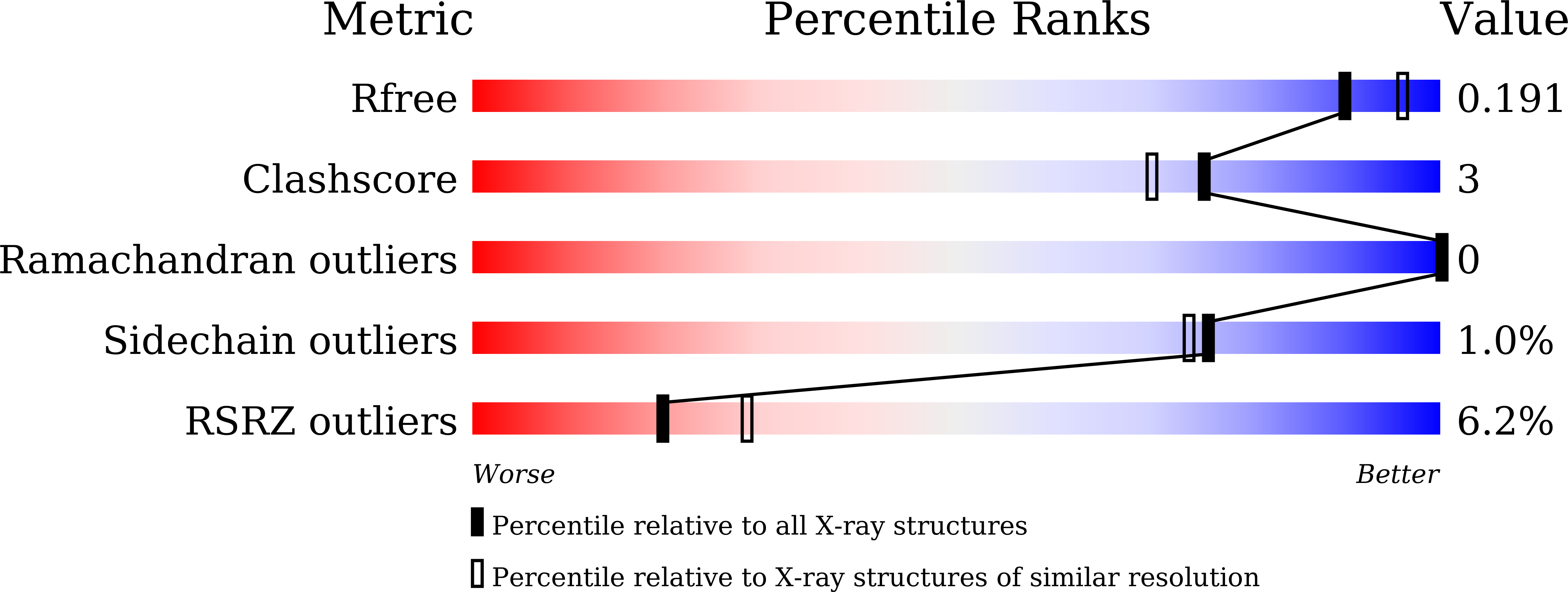
Structural insights into the dimer-tetramer transition of FabI from Bacillus anthracis
Kim, H.T., Kim, S., Na, B.K., Chung, J., Hwang, E., Hwang, K.Y.(2017) Biochem Biophys Res Commun 493: 28-33
- PubMed: 28935372
- DOI: https://doi.org/10.1016/j.bbrc.2017.09.084
- Primary Citation of Related Structures:
5YCR, 5YCS, 5YCV, 5YCX - PubMed Abstract:
Enoyl-ACP reductase (ENR, also known as FabI) has received considerable interest as an anti-bacterial target due to its essentiality in fatty acid synthesis. All the FabI structures reported to date, regardless of the organism, are composed of homo-tetramers, except for two structures: Bacillus cereus and Staphylococcus aureus FabI (bcFabI and saFabI, respectively), which have been reported as dimers. However, the reason for the existence of the dimeric form in these organisms and the biological meaning of dimeric and tetrameric forms of FabI are ambiguous. Herein, we report the high-resolution crystal structure of a dimeric form of Bacillus anthracis FabI (baFabI) and the crystal structures of tetrameric forms of baFabI in the apo state and in complex with NAD + and with NAD + -triclosan, at 1.7 Å, 1.85 Å, 1.96 Å, and 1.95 Å, respectively. Interestingly, we found that baFabI with a His 6 -tag at its C-terminus exists as a dimer, whereas untagged-baFabI exists as a tetramer. The His 6 -tag may block the dimer-tetramer transition, since baFabI has relatively short-length amino acids ( 255 LG 256 ) after the 3 10 -helix η7 compared to those of FabI of other organisms. The dimeric form of baFabI is catalytically inactive, because the α-helix α5 occupies the NADH-binding site. During the process of dimer-tetramer transition, this α5 helix rotates about 55° toward the tetramer interface and the active site is established. Therefore, tetramerization of baFabI is required for cofactor binding and catalytic activity.
Organizational Affiliation:
Crystalgenomics, Inc., 5F, Tower A, Korea Bio Park 700, Daewangpangyo-ro, Bundang-gu, Seongnam-si, Gyeonggi-do 13524, South Korea; Division of Biotechnology, College of Life Sciences and Biotechnology, Korea University, 145 Anam-ro, Seongbuk-gu, Seoul 02841, South Korea.