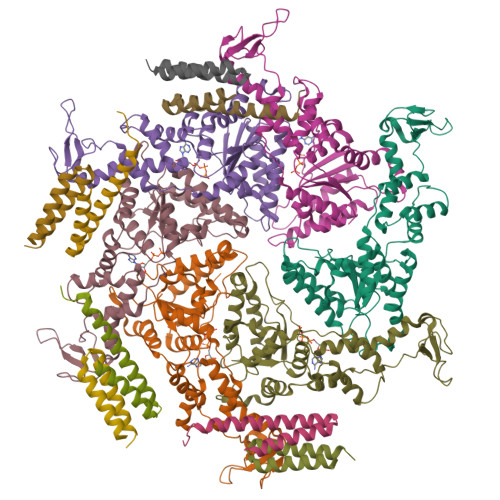
Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Sun, S., Li, L., Yang, F., Wang, X., Fan, F., Yang, M., Chen, C., Li, X., Wang, H.W., Sui, S.F.(2017) Nat Commun 8: 16064-16064
- PubMed: 28714467
- DOI: https://doi.org/10.1038/ncomms16064
- Primary Citation of Related Structures:
5XMI, 5XMK - PubMed Abstract:
The cellular ESCRT-III (endosomal sorting complex required for transport-III) and Vps4 (vacuolar protein sorting 4) comprise a common machinery that mediates a variety of membrane remodelling events. Vps4 is essential for the machinery function by using the energy from ATP hydrolysis to disassemble the ESCRT-III polymer into individual proteins. Here, we report the structures of the ATP-bound Vps4 E233Q hexamer and its complex with the cofactor Vta1 (vps twenty associated 1) at resolutions of 3.9 and 4.2 Å, respectively, determined by electron cryo-microscopy. Six Vps4 E233Q subunits in both assemblies exhibit a spiral-shaped ring-like arrangement. Locating at the periphery of the hexameric ring, Vta1 dimer bridges two adjacent Vps4 subunits by two different interaction modes to promote the formation of the active Vps4 hexamer during ESCRT-III filament disassembly. The structural findings, together with the structure-guided biochemical and single-molecule analyses, provide important insights into the process of the ESCRT-III polymer disassembly by Vps4.
Organizational Affiliation:
State Key Laboratory of Membrane Biology, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.